Improved Astaxanthin Production with Corynebacterium glutamicum by Application of a Membrane Fusion Protein
Abstract
:1. Introduction
2. Results
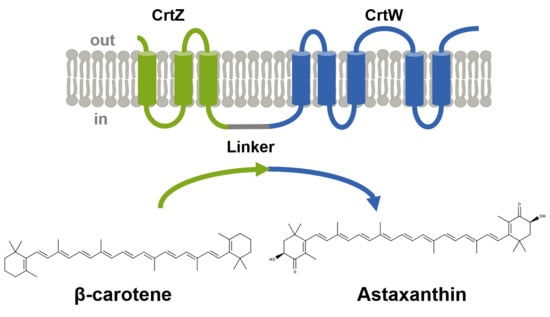
2.1. Fusion of the Membrane-bound Proteins β-carotene Hydroxylase and β-carotene Hydroxylase Increased Astaxanthin Production under High Glucose Concentration
2.2. Overproduction of Cytosolic Carotenoid Enzymes Increased Astaxanthin Production under High Glucose Concentration
2.3. Co-utilization of Glucose and Potassium Acetate Improved Astaxanthin Production in Minimal Medium
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Growth Conditions
4.2. Cloning of Expression Vectors
4.3. Construction of pSH1- and pECXT99A-based Expression Vectors
4.4. Carotenoid Quantification
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- GrandViewResearch. Astaxanthin Market Analysis By Source (Natural [Yeast, Krill/Shrimp, Microalgae] And Synthetic), By Product (Dried Biomass/Powder, Oil, Soft gels, Liquid), By Application, And Segment Forecasts, 2018–2025. 2017. Available online: https://www.grandviewresearch.com/industry-analysis/global-astaxanthin-market (accessed on 1 August 2019).
- GrandViewResearch. Compound Feed Market Analysis, Market Size, Application Analysis, Regional Outlook, Competitive Strategies, And Segment Forecasts, 2016–2024. 2016. Available online: https://www.grandviewresearch.com/industry-analysis/compound-feed-market (accessed on 1 August 2019).
- Wendisch, V.F.; Mindt, M.; Perez-Garcia, F. Biotechnological production of mono- and diamines using bacteria: Recent progress, applications, and perspectives. Appl. Microbiol. Biotechnol. 2018, 102, 3583–3594. [Google Scholar] [CrossRef] [PubMed]
- Wendisch, V.F. L-Lysine. In Industrial Biotechnology; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2017; pp. 361–390. [Google Scholar] [CrossRef]
- Hirasawa, T.; Wachi, M. Glutamate Fermentation-2: Mechanism of L-Glutamate Overproduction in Corynebacterium glutamicum. Adv. Biochem. Eng. Biotechnol. 2017, 159, 57–72. [Google Scholar] [CrossRef] [PubMed]
- Heider, S.A.; Wolf, N.; Hofemeier, A.; Peters-Wendisch, P.; Wendisch, V.F. Optimization of the IPP precursor supply for the production of lycopene, decaprenoxanthin and astaxanthin by Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 2014, 2, 28. [Google Scholar] [CrossRef] [PubMed]
- Heider, S.A.; Peters-Wendisch, P.; Wendisch, V.F. Carotenoid biosynthesis and overproduction in Corynebacterium glutamicum. BMC Microbiol. 2012, 12, 198. [Google Scholar] [CrossRef] [PubMed]
- Krubasik, P.; Takaichi, S.; Maoka, T.; Kobayashi, M.; Masamoto, K.; Sandmann, G. Detailed biosynthetic pathway to decaprenoxanthin diglucoside in Corynebacterium glutamicum and identification of novel intermediates. Arch. Microbiol. 2001, 176, 217–223. [Google Scholar] [CrossRef]
- Krubasik, P.; Kobayashi, M.; Sandmann, G. Expression and functional analysis of a gene cluster involved in the synthesis of decaprenoxanthin reveals the mechanisms for C50 carotenoid formation. Eur. J. Biochem. 2001, 268, 3702–3708. [Google Scholar] [CrossRef]
- Henke, N.A.; Heider, S.A.E.; Hannibal, S.; Wendisch, V.F.; Peters-Wendisch, P. Isoprenoid Pyrophosphate-Dependent Transcriptional Regulation of Carotenogenesis in Corynebacterium glutamicum. Front. Microbiol. 2017, 8, 633. [Google Scholar] [CrossRef]
- Taniguchi, H.; Henke, N.A.; Heider, S.A.E.; Wendisch, V.F. Overexpression of the primary sigma factor gene sigA improved carotenoid production by Corynebacterium glutamicum: Application to production of β-carotene and the non-native linear C50 carotenoid bisanhydrobacterioruberin. Metab. Eng. Commun. 2017, 4, 1–11. [Google Scholar] [CrossRef]
- Heider, S.A.; Peters-Wendisch, P.; Netzer, R.; Stafnes, M.; Brautaset, T.; Wendisch, V.F. Production and glucosylation of C50 and C40 carotenoids by metabolically engineered Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2014, 98, 1223–1235. [Google Scholar] [CrossRef]
- Henke, N.A.; Heider, S.A.; Peters-Wendisch, P.; Wendisch, V.F. Production of the Marine Carotenoid Astaxanthin by Metabolically Engineered Corynebacterium glutamicum. Mar. Drugs 2016, 14, 124. [Google Scholar] [CrossRef]
- Ye, L.; Zhu, X.; Wu, T.; Wang, W.; Zhao, D.; Bi, C.; Zhang, X. Optimizing the localization of astaxanthin enzymes for improved productivity. Biotechnol. Biofuels 2018, 11, 278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camagna, M.; Grundmann, A.; Bar, C.; Koschmieder, J.; Beyer, P.; Welsch, R. Enzyme Fusion Removes Competition for Geranylgeranyl Diphosphate in Carotenogenesis. Plant Physiol. 2019, 179, 1013–1027. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yan, P.; Liu, X.; Wang, Z.; Tang, Y.J.; Chen, T.; Zhao, X. Combinatorial expression of different beta-carotene hydroxylases and ketolases in Escherichia coli for increased astaxanthin production. J. Ind. Microbiol. Biotechnol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef] [PubMed]
- Gerstmeir, R.; Wendisch, V.F.; Schnicke, S.; Ruan, H.; Farwick, M.; Reinscheid, D.; Eikmanns, B.J. Acetate metabolism and its regulation in Corynebacterium glutamicum. J. Biotechnol. 2003, 104, 99–122. [Google Scholar] [CrossRef]
- Knoll, A.; Bartsch, S.; Husemann, B.; Engel, P.; Schroer, K.; Ribeiro, B.; Stockmann, C.; Seletzky, J.; Buchs, J. High cell density cultivation of recombinant yeasts and bacteria under non-pressurized and pressurized conditions in stirred tank bioreactors. J. Biotechnol. 2007, 132, 167–179. [Google Scholar] [CrossRef]
- Riesenberg, D.; Guthke, R. High-cell-density cultivation of microorganisms. Appl. Microbiol. Biotechnol. 1999, 51, 422–430. [Google Scholar] [CrossRef]
- Sasaki, Y.; Eng, T.; Herbert, R.A.; Trinh, J.; Chen, Y.; Rodriguez, A.; Gladden, J.; Simmons, B.A.; Petzold, C.J.; Mukhopadhyay, A. Engineering Corynebacterium glutamicum to produce the biogasoline isopentenol from plant biomass hydrolysates. Biotechnol. Biofuels 2019, 12, 41. [Google Scholar] [CrossRef]
- Henke, N.A.; Wiebe, D.; Pérez-García, F.; Peters-Wendisch, P.; Wendisch, V.F. Coproduction of cell-bound and secreted value-added compounds: Simultaneous production of carotenoids and amino acids by Corynebacterium glutamicum. Bioresour. Technol. 2018, 722–752. [Google Scholar] [CrossRef]
- Pompon, D.; Garcia-Alles, L.; Truan, G. Nanotechnology for Synthetic Biology: Crossroads Throughout Spatial Confinement. In Nanotechnology in Agriculture and Food Science; Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2017; pp. 209–234. [Google Scholar] [CrossRef]
- Schmitt, D.L.; An, S. Spatial Organization of Metabolic Enzyme Complexes in Cells. Biochemistry 2017, 56, 3184–3196. [Google Scholar] [CrossRef] [Green Version]
- Srere, P.A. The metabolon. Trends Biochem. Sci. 1985, 10, 109–110. [Google Scholar] [CrossRef]
- Rabeharindranto, H.; Castano-Cerezo, S.; Lautier, T.; Garcia-Alles, L.F.; Treitz, C.; Tholey, A.; Truan, G. Enzyme-fusion strategies for redirecting and improving carotenoid synthesis in S. cerevisiae. Metab. Eng. Commun. 2019, 8, e00086. [Google Scholar] [CrossRef] [PubMed]
- Barredo, J.L.; Garcia-Estrada, C.; Kosalkova, K.; Barreiro, C. Biosynthesis of Astaxanthin as a Main Carotenoid in the Heterobasidiomycetous Yeast Xanthophyllomyces dendrorhous. J. Fungi 2017, 3, 44. [Google Scholar] [CrossRef] [PubMed]
- Martín, J.F.; Gudiña, E.; Barredo, J.L. Conversion of beta-carotene into astaxanthin: Two separate enzymes or a bifunctional hydroxylase-ketolase protein? Microb. Cell Fact. 2008, 7, 3. [Google Scholar] [CrossRef]
- Nogueira, M.; Enfissi, E.M.A.; Welsch, R.; Beyer, P.; Zurbriggen, M.D.; Fraser, P.D. Construction of a fusion enzyme for astaxanthin formation and its characterisation in microbial and plant hosts: A new tool for engineering ketocarotenoids. Metab. Eng. 2019, 52, 243–252. [Google Scholar] [CrossRef]
- Albertsen, L.; Chen, Y.; Bach, L.S.; Rattleff, S.; Maury, J.; Brix, S.; Nielsen, J.; Mortensen, U.H. Diversion of flux toward sesquiterpene production in Saccharomyces cerevisiae by fusion of host and heterologous enzymes. Appl. Environ. Microbiol. 2011, 77, 1033–1040. [Google Scholar] [CrossRef]
- Chen, X.; Zaro, J.L.; Shen, W.-C. Fusion protein linkers: Property, design and functionality. Adv. Drug Deliv. Rev. 2013, 65, 1357–1369. [Google Scholar] [CrossRef]
- George, R.A.; Heringa, J. An analysis of protein domain linkers: Their classification and role in protein folding. Protein Eng. 2002, 15, 871–879. [Google Scholar] [CrossRef]
- Park, S.Y.; Binkley, R.M.; Kim, W.J.; Lee, M.H.; Lee, S.Y. Metabolic engineering of Escherichia coli for high-level astaxanthin production with high productivity. Metab. Eng. 2018, 49, 105–115. [Google Scholar] [CrossRef]
- Wendisch, V.F.; de Graaf, A.A.; Sahm, H.; Eikmanns, B.J. Quantitative determination of metabolic fluxes during coutilization of two carbon sources: Comparative analyses with Corynebacterium glutamicum during growth on acetate and/or glucose. J. Bacteriol. 2000, 182, 3088–3096. [Google Scholar] [CrossRef]
- Misawa, N. Carotenoid beta-ring hydroxylase and ketolase from marine bacteria-promiscuous enzymes for synthesizing functional xanthophylls. Mar. Drugs 2011, 9, 757–771. [Google Scholar] [CrossRef] [PubMed]
- Fraser, P.D.; Miura, Y.; Misawa, N. In vitro characterization of astaxanthin biosynthetic enzymes. J. Biol. Chem. 1997, 272, 6128–6135. [Google Scholar] [CrossRef] [PubMed]
- Durnin, G.; Clomburg, J.; Yeates, Z.; Alvarez, P.J.; Zygourakis, K.; Campbell, P.; Gonzalez, R. Understanding and harnessing the microaerobic metabolism of glycerol in Escherichia coli. Biotechnol. Bioeng. 2009, 103, 148–161. [Google Scholar] [CrossRef] [PubMed]
- Liebl, W. Corynebacterium taxonomy. In Handbook of Corynebacterium glutamicum; Eggeling, L., Bott, M., Eds.; CRC Press: Boca Raton, FL, USA, 2005; pp. 9–34. [Google Scholar]
- Baumgart, M.; Unthan, S.; Ruckert, C.; Sivalingam, J.; Grunberger, A.; Kalinowski, J.; Bott, M.; Noack, S.; Frunzke, J. Construction of a prophage-free variant of Corynebacterium glutamicum ATCC 13032 for use as a platform strain for basic research and industrial biotechnology. Appl. Environ. Microbiol. 2013, 79, 6006–6015. [Google Scholar] [CrossRef]
- Abe, S.; Takayarna, K.; Kinoshita, S. Taxonomical studies on glutamic acid producing bacteria. J. Gen. Appl. Microbiol. 1967, 13, 279–301. [Google Scholar] [CrossRef]
- Hanahan, D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983, 166, 557–580. [Google Scholar] [CrossRef]
- Misawa, N.; Nakagawa, M.; Kobayashi, K.; Yamano, S.; Izawa, Y.; Nakamura, K.; Harashima, K. Elucidation of the Erwinia uredovora carotenoid biosynthetic pathway by functional analysis of gene products expressed in Escherichia coli. J. Bacteriol. 1990, 172, 6704–6712. [Google Scholar] [CrossRef]
- Cho, J.C.; Giovannoni, S.J. Fulvimarina pelagi gen. nov., sp. nov., a marine bacterium that forms a deep evolutionary lineage of descent in the order “Rhizobiales”. Int. J. Syst. Evol. Microbiol. 2003, 53, 1853–1859. [Google Scholar] [CrossRef]
- Kirchner, O.; Tauch, A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 2003, 104, 287–299. [Google Scholar] [CrossRef]
- Gibson, D.G.; Young, L.; Chuang, R.Y.; Venter, J.C.; Hutchison, C.A., 3rd; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef]
- van der Rest, M.E.; Lange, C.; Molenaar, D. A heat shock following electroporation induces highly efficient transformation of Corynebacterium glutamicum with xenogeneic plasmid DNA. Appl. Microbiol. Biot. 1999, 52, 541–545. [Google Scholar] [CrossRef] [PubMed]





| Strain | Characteristics | Reference |
|---|---|---|
| C. glutamicum strains | ||
| WT | Wild type, ATCC 13032 | [40] |
| MB001 | prophage cured, genome reduced ATCC 13032 | [39] |
| BETA4 | MB001 derivative with deletion of crtYEb (cg0717-0719) and crtR (cg0725) and integration of Ptuf-dxs, Ptuf-crtEBI, Ptuf-crtYPa | [13] |
| ASTA1 | BETA4 derivative with pSH1-crtWFp and pECXT99A-crtZFp | [13] |
| ASTA* | BETA4 derivative with pSH1-crtZ~WFp | This work |
| Other strains | ||
| E. coli DH5α | F-thi-1 endA1 hsdr17(r-, m-) supE44 ΔlacU169 (Φ80lacZΔM15) recA1 gyrA96 | [41] |
| Pantoea ananatis | Wild type, ATCC 33244, DSM 17873, Z96081 | [42] |
| Fulvimarina pelagi | Wild type, ATCC BAA-666, DSM 15513, AY178860 | [43] |
| Plasmids | ||
| pECXT99A (pECXT) | TetR, PtrclacIq, pGA1 oriVCg, C. glutamicum/E. coli expression shuttle vector | [44] |
| pECXT-idi | pECXT99A derivative for IPTG-inducible expression of idi (cg2531) | This work |
| pECXT-idsA | pECXT99A derivative for IPTG-inducible expression of idsA (cg2384) | This work |
| pECXT- crtBI | pECXT99A derivative for IPTG-inducible expression of crtBI (cg0721/0720) | This work |
| pECXT-idi-idsA | pECXT99A derivative for IPTG-inducible expression of idi (cg2531) and idsA (cg2384) | This work |
| pECXT-idsA-crtBI | pECXT99A derivative for IPTG-inducible expression of idsA (cg2384) and crtBI (cg0721/0720) | This work |
| pECXT-idi-idsA-crtBI | pECXT99A derivative for IPTG-inducible expression of idi (cg2531), idsA (cg2384) and crtBI (cg0721/0720) | This work |
| pSH1 | KmR, Ptuf, pHM519 oriVCg, C. glutamicum/E. coli expression shuttle vector | [13] |
| pSH1-crtW-crtZ | pSH1 derivative for constitutive expression of the artificial operon comprising crtW and crtZ from F. pelagi | This work |
| pSH1-crtW~Z | pSH1 derivative for constitutive expression of crtW~Z encoding for a fusion protein comprising CrtW and CrtZ from F. pelagi | This work |
| pSH1-crtZ-crtW | pSH1 derivative for constitutive expression of the artificial operon comprising crtZ and crtW from F. pelagi | This work |
| pSH1-crtZ~W | pSH1 derivative for constitutive expression of crtZ~W encoding for a fusion protein comprising CrtZ and CrtW from F. pelagi | This work |
| Oligo-nucleotide | Target | Sequence (5’ → 3’) |
|---|---|---|
| FpW1 | Wfw1 | CATGCCTGCAGGTCGACTCTAGAGGAAAGGAGGCCCTTCAGATGACCCTCAGCCCAACCTC |
| HN05 | Wrv1 | GTTCGTGTGGCAGTTTTAGGACTGGCGAGTATGCG |
| HN06 | Zfw1 | AACTGCCACACGAACGAAAGGAGGCCCTTCAGATGACGATCTGGACTCTCTACTAC |
| HA35 | Zrv1 | ATTCGAGCTCGGTACCCGGGGATCTTACCGAACCGGCGCGT |
| HA47 | Wrv-L | CGGAACCGCCACCGCCGGACTGGCGAGTATG |
| HA48 | L-Zfw | GGCGGTGGCGGTTCCGGCGGTCCAGGTTCCACGATCTGGACTCTCTACTAC |
| HA34 | Zfw2 | CATGCCTGCAGGTCGACTCTAGAGGAAAGGAGGCCCTTCAGATGACGATCTGGACTCTCTACTAC |
| HA45 | Zrv2 | GTTCGTGTGGCAGTTTTACCGAACCGGCGCGT |
| HA46 | Wfw2 | AACTGCCACACGAACGAAAGGAGGCCCTTCAGATGACCCTCAGCCCAACCTC |
| FpW4 | Wrv2 | ATTCGAGCTCGGTACCCGGGGATCTTAGGACTGGCGAGTATGCG |
| HA49 | Zrv-L | CGGAACCGCCACCGCCCCGAACCGGCGCGT |
| HA50 | L-Wfw | GGCGGTGGCGGTTCCGGCGGTCCAGGTTCCACCCTCAGCCCAACCTC |
| HA67 | Idifw | ATGGAATTCGAGCTCGGTACCCGGGGAAAGGAGGCCCTTCAGATGTCTAAGCTTAGGGGCATGAC |
| HA68 | Idirv | GCATGCCTGCAGGTCGACTCTAGAGGATCTTACTCTGCGTCAAACGCTTC |
| HA69 | CrtBIfw | ATGGAATTCGAGCTCGGTACCCGGGGAAAGGAGGCCCTTCAGATGACACACCAAAATTCGCC |
| HA70 | CrtBIrv | GCATGCCTGCAGGTCGACTCTAGAGGATCTTAATGATCGTATGAGGTCTTTTGAG |
| NH56 | idsArv | GCATGCCTGCAGGTCGACTCTAGAGGATCTTACATCCGACGTTCGGTTGA |
| NH55 | idsAfw | ATGGAATTCGAGCTCGGTACCCGGGGAAAGGAGGCCCTTCAGATGAGCAGTTTCGATGCCCA |
| NH57 | idsA-rv-L | GTTCGTGTGGCAGTTTTACATCCGACGTTCGGTTGA |
| HA71 | L-CrtBIfw | AACTGCCACACGAACGAAAGGAGGCCCTTCAGATGACACACCAAAATTCGCC |
| PD5 | ACCGGCTCCAGATTTATCAG | |
| 582 | ATCTTCTCTCATCCTCCA | |
| pEC-fw | AATACGCAAACCGCCTCTCC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Henke, N.A.; Wendisch, V.F. Improved Astaxanthin Production with Corynebacterium glutamicum by Application of a Membrane Fusion Protein. Mar. Drugs 2019, 17, 621. https://doi.org/10.3390/md17110621
Henke NA, Wendisch VF. Improved Astaxanthin Production with Corynebacterium glutamicum by Application of a Membrane Fusion Protein. Marine Drugs. 2019; 17(11):621. https://doi.org/10.3390/md17110621
Chicago/Turabian StyleHenke, Nadja A., and Volker F. Wendisch. 2019. "Improved Astaxanthin Production with Corynebacterium glutamicum by Application of a Membrane Fusion Protein" Marine Drugs 17, no. 11: 621. https://doi.org/10.3390/md17110621