Chlorophyll Derivatives from Marine Cyanobacteria with Lipid-Reducing Activities
Abstract
:1. Introduction
2. Results
2.1. Isolation of Compound 1
2.2. Isolation and Sctructure Elucidation of Compound 2
2.3. Lipid-Reducing Activity of 1 and 2, but not of Chlorophyll a and b
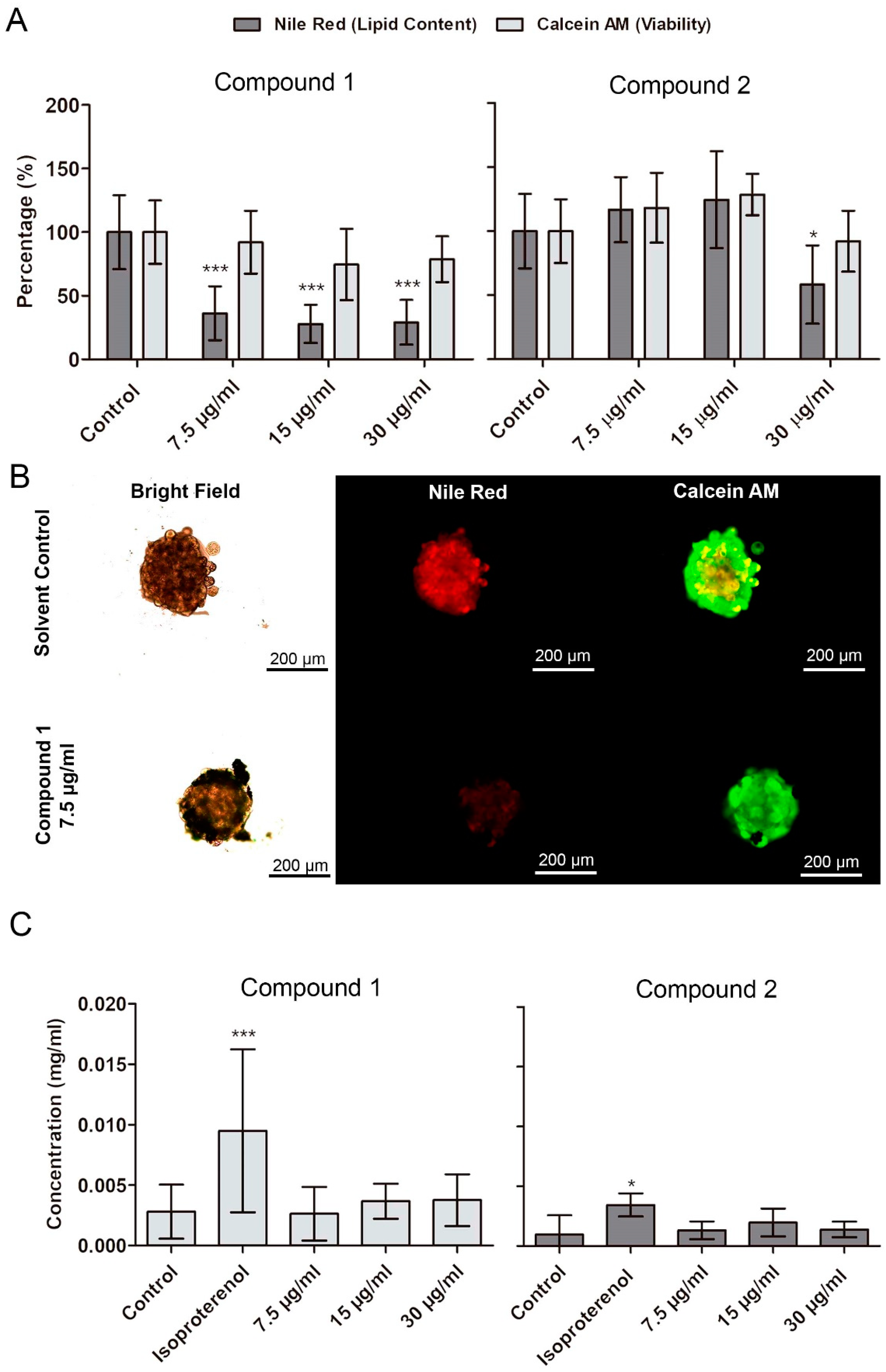
2.4. Confirmation of Lipid-Reducing Activity in Differentiated 3T3-L1 Spheroids, and Analysis of Lipolysis
2.5. qPCR Indicates PPARγ for 1
2.6. Quantification in Different Source Material of 1
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Cyanobacterial Growth, Extraction and Fractionation
4.3. Compound 1 Isolation and Structure Elucidation
4.4. Compound 2 Isolation and Structure Elucidation
4.5. Determination of 132-hydroxy-pheophytin a by LC–MS
4.6. Zebrafish Nile Red Fat Metabolism Assay
4.7. Differentiated Murine Preadipocytes Grown as Spheroids
4.8. Lipolysis
4.9. Real-Time PCR
4.10. Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fu, C.; Jiang, Y.; Guo, J.; Su, Z. Natural products with anti-obesity effects and different mechanisms of action. J. Agric. Food Chem. 2016, 64, 9571–9585. [Google Scholar] [CrossRef] [PubMed]
- Castro, M.; Preto, M.; Vasconcelos, V.; Urbatzka, R. Obesity: The metabolic disease, advances on drug discovery and natural product research. Curr. Top. Med. Chem. 2016, 16, 2577–2604. [Google Scholar] [CrossRef] [PubMed]
- Fox, C.; Massaro, J.; Hoffmann, U.; Pou, K.; Maurovich-Horvat, P.; Liu, C.; Vasan, R.; Murabito, J.; Meigs, J.; Cupples, L.; et al. Abdominal visceral and subcutaneous adipose tissue compartments: Association Pereira DCR. Circulation 2007, 116, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.G.; Park, C.-Y. Anti-Obesity Drugs: A Review about Their Effects and Safety. Diabetes Obes. Metab. 2012, 36, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Daneschvar, H.L.; Aronson, M.D.; Smetana, G.W. FDA-approved anti-obesity drugs in the United States. Am. J. Med. 2016, 129, e1–e6. [Google Scholar] [CrossRef]
- Inuzuka, T.; Yamamoto, K.; Iwasaki, A.; Ohno, O.; Suenaga, K.; Kawazoe, Y.; Uemura, D. An inhibitor of the adipogenic differentiation of 3T3-L1 cells, yoshinone A, and its analogs, isolated from the marine cyanobacterium Leptolyngbya sp. Tetrahedron Lett. 2014, 55, 6711–6714. [Google Scholar] [CrossRef]
- Blunt, J.W.; Carrol, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Princep, M.R. Marine natural products. Nat. Prod. Rep. 2018, 35, 8–53. [Google Scholar] [CrossRef] [Green Version]
- Luzzatto-Knaan, T.; Garg, N.; Wang, M.; Glukhov, E.; Peng, Y.; Ackemann, G.; Amir, A.; Duggan, B.M.; Ryazanov, S.; Gerwick, L.; et al. Digitizing mass spectrometry data to explore the chemical diversity and distribution of marine cyanobacteria and algae. eLife 2017, 6, e24214. [Google Scholar] [CrossRef]
- Brito, A.; Gaifem, J.; Ramos, V.; Glukhov, E.; Dorrestein, P.C.; Gerwick, W.H.; Vasconcelos, V.M.; Mendes, M.V.; Tamagnini, P. Bioprospecting portuguese atlantic coast cyanobacteria for bioactive secondary metabolites reveals untapped chemodiversity. Algal Res. 2015, 9, 218–226. [Google Scholar] [CrossRef]
- Sharma, N.K.; Tiwari, S.P.; Tripathi, K.; Rai, A.K. Sustainability and cyanobacteria (blue-green algae): Facts and challenges. J. Appl. Phycol. 2011, 23, 1059–1081. [Google Scholar] [CrossRef]
- Ramos, V.; Morais, J.; Castelo-Branco, R.; Pinheiro, A.; Martins, J.; Regueiras, A.; Pereira, A.L.; Lopes, V.R.; Frazão, B.; Gomes, D.; et al. Cyanobacterial diversity held in microbial biological resource centers as a biotechnological asset: The case study of the newly established LEGE culture collection. J. Appl. Phycol. 2018, 30, 1437–1451. [Google Scholar] [CrossRef]
- Leão, P.N.; Costa, M.; Ramos, V.; Pereira, A.R.; Fernandes, V.C.; Domingues, V.F.; Gerwick, W.H.; Vasconcelos, V.M.; Martins, R. Antitumor Activity of Hierridin B, a Cyanobacterial Secondary Metabolite Found in both Filamentous and Unicellular Marine Strains. PLoS ONE 2013, 8, e69562. [Google Scholar] [CrossRef]
- Leão, P.N.; Nakamura, H.; Costa, M.; Pereira, A.R.; Martins, R.; Vasconcelos, V.; Gerwick, W.H.; Balskus, E.P. Biosynthesis-assisted structural elucidation of the bartolosides, chlorinated aromatic glycolipids from cyanobacteria. Angew. Chem. Int. Ed. Engl. 2015, 54, 11063–11067. [Google Scholar] [CrossRef] [PubMed]
- Afonso, T.B.; Costa, M.S.; Rezende de Castro, R.; Freitas, S.; Silva, A.; Schneider, M.P.C.; Martins, R.; Leão, P.N. Bartolosides E-K from a marine coccoid cyanobacterium. Nat. Prod. 2016, 79, 2504–2513. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, T.; Lemos, F.; Preto, M.; Azevedo, J.; Sousa, M.L.; Leão, P.N.; Campos, A.; Linder, S.; Vitorino, R.; Vasconcelos, V.; et al. Cytotoxicity of portoamides in human cancer cells and analysis of the molecular mechanisms of action. PLoS ONE 2017, 12, e0188817. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.; Yang, E.; Neelkantan, N.; Mikhaylova, A.; Arnold, R.; Poudel, M.K.; Stewart, A.M.; Kalueff, A.V. Developing ‘integrative’ zebrafish models of behavioral and metabolic disorders. Behav. Brain Res. 2013, 256, 172–187. [Google Scholar] [CrossRef]
- Oka, T.; Nishimura, Y.; Zang, L.; Hirano, M.; Shimada, Y.; Wang, Z.; Umemoto, N.; Kuroyanagi, J.; Nishimura, N.; Tanaka, T. Diet-induced obesity in zebrafish shares common pathophysiological pathways with mammalian obesity. BMC Physiol. 2010, 10, 21. [Google Scholar] [CrossRef] [PubMed]
- Jones, K.S.; Alimov, A.P.; Rilo, H.L.; Jandacek, R.J.; Woollett, L.A.; Penberthy, W.T. A high throughput live transparent animal bioassay to identify non-toxic small molecules or genes that regulate vertebrate fat metabolism for obesity drug development. Nutr. Metab. 2008, 5, 1–11. [Google Scholar] [CrossRef]
- Jerz, G.; Arrey, T.N.; Wray, V.; Du, Q.; Winterhalter, P. Structural characterization of 132-hydroxy-(132-S)-phaeophytin-a from leaves and stems of Amaranthus tricolor isolated by high-speed countercurrent chromatography. Innov. Food Sci. Emerg. Technol. 2007, 8, 413–418. [Google Scholar] [CrossRef]
- Yaacob, N.S.; Yankuzo, H.M.; Devaraj, S.; Wong, J.K.M.; Lai, C.-S. Anti-Tumor Action, Clinical biochemistry profile and phytochemical constituents of a pharmacologically active fraction of S. crispus in NMU-Induced Rat Mammary Tumour Model. PLoS ONE 2015, 10, e0126426. [Google Scholar] [CrossRef]
- Lee, T.H.; Lu, C.K.; Kuo, Y.H.; Lo, J.M.; Lee, C.K. Unexpected novel pheophytin peroxides from the leaves of Biden Pilosa. Helv. Chim. Acta 2008, 91, 79–84. [Google Scholar] [CrossRef]
- Li, H.; Li, L.; Zheng, Q.; Kuroda, C.; Wang, Q. Phaeophytin analogues from Ligularia knorringiana. Molecules 2012, 17, 5219–5224. [Google Scholar] [CrossRef] [PubMed]
- Masuda, T.; Fujita, Y. Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 2008, 7, 1131–1149. [Google Scholar] [CrossRef]
- Matsuo, A.; Ono, K.; Hamasaki, K.; Nozaki, H. Phaeophytins from a cell suspension culture of the liverwort Plagiochila ovalifolia. Phytochemistry 1996, 42, 427–430. [Google Scholar] [CrossRef]
- Nakatani, Y.; Ourisson, G.; Beck, J.P. Chemistry and biochemistry of chinese drugs. VII. Cytostatic pheophytins from silkworm excreta, and derived photocytotoxic pheophorbides. Chem. Pharm. Bull. 1981, 29, 2261–2269. [Google Scholar] [CrossRef] [PubMed]
- Desarzens, S.; Liao, W.H.; Mammi, C.; Caprio, M.; Faresse, N. Hsp90 blockers inhibit adipocyte differentiation and fat mass accumulation. PLoS ONE 2014, 9, e94127. [Google Scholar] [CrossRef] [PubMed]
- Aronoff, S. The chlorophylls—An introductory survey. In The Chlorophylls; Vernon, L.P., Seely, G.R., Eds.; Academic Press: New York, NY, USA, 1996; pp. 3–20. [Google Scholar]
- Scheer, H. Structure and occurrence of chlorophylls. In Chlorophylls; Scheer, H., Ed.; CRC Press: Boca Raton, FL, USA, 1991; pp. 3–30. [Google Scholar]
- Tanaka, R.; Tanaka, A. Tetrapyrrole biosynthesis in higher plants. Annu. Rev. Plant Biol. 2007, 58, 321–346. [Google Scholar] [CrossRef]
- Vavilin, D.V.; Vermaas, W.F. Regulation of the tetrapyrrole biosynthetic pathway leading to heme and chlorophyll in plants and cyanobacteria. Physiol. Plant 2002, 115, 9–24. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.-R.; Park, J.; Jung, S.K.; Chang, Y.H. Synthesis, characterization, and functional properties of chlorophylls, pheophytins, and Zn-pheophytins. Food Chem. 2018, 245, 943–950. [Google Scholar] [CrossRef]
- Subramoniam, A.; Asha, V.V.; Nair, S.A.; Sasidharan, S.P.; Sureshkumar, P.K.; Rajendran, K.N.; Karunagaran, D.; Ramalingam, K. Chlorophyll revisited: Anti-inflammatory activities of chlorophyll a and inhibition of expression of TNF-α gene by the same. Inflammation 2012, 35, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Ferruzzi, M.; Böhm, V.; Courtney, P.; Schwartz, S. Antioxidant and antimutagenic activity of dietary chlorophyll derivatives determined by radical scavenging and bacterial reverse mutagenesis assays. J. Food Sci. Technol. 2002, 67, 2589–2595. [Google Scholar] [CrossRef]
- Zhao, Y.; Wang, X.; Wang, H.; Liu, T.; Xin, Z. Two new noroleanane-type triterpene saponins from the methanol extract of Salicornia herbacea. Food Chem. 2014, 151, 101–109. [Google Scholar] [CrossRef]
- Wang, S.-Y.; Tseng, C.-P.; Tsai, K.-C.; Lin, C.-F.; Wen, C.-Y.; Tsay, H.-S.; Sakamoto, N.; Tseng, C.-H.; Cheng, J.-C. Bioactivity-guided screening identifies pheophytin a as a potent anti-hepatitis C virus compound from Lonicera hypoglauca Miq. Biochem. Biophys. Res. Commun. 2009, 385, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Gomes, R.A.; Teles, Y.C.; Pereira, F.D.; Rodrigues, L.A.; Lima, E.D.; Agra, M.D.; Souza, M.D. Phytoconstituents from Sidastrum micranthum (A. St.-Hil.) Fryxell (Malvaceae) and antimicrobial activity of pheophytin a. Braz. J. Pharm. Sci. 2015, 51, 861–867. [Google Scholar] [CrossRef]
- Ina, A.; Hayashi, K.-I.; Nozaki, H.; Kamei, Y. Pheophytin A, a low molecular weight compound found in the marine brown alga Sargassum fulvellum, promotes the differentiation of PC12 cells. Int. J. Dev. Neurosci. 2007, 25, 63–68. [Google Scholar] [CrossRef]
- Sharifuddin, Y.; Chin, Y.-X.; Lim, P.-E.; Phang, S.-M. Potential bioactive compounds from seaweed for diabetes management. Mar. Drugs 2015, 13, 5447–5491. [Google Scholar] [CrossRef]
- Semaan, D.G.; Igoli, J.O.; Young, L.; Gray, A.I.; Rowan, E.G.; Marrero, E. In vitro anti-diabetic effect of flavonoids and pheophytins from Allophylus cominia Sw. on the glucose uptake assays by HepG2, L6, 3T3-L1 and fat accumulation in 3T3-L1 adipocytes. J. Ethnopharmacol. 2018, 216, 8–17. [Google Scholar] [CrossRef]
- Cheng, H.H.; Wang, H.K.; Ito, J.; Bastow, K.F.; Tachibana, Y.; Nakanishi, Y.; Xu, Z.; Luo, T.Y.; Lee, K.H. Cytotoxic pheophorbide-related compounds from Clerodendrum calamitosum and C. cyrtophyllum. J. Nat. Prod. 2001, 64, 915–919. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; He, J.; Tong, X.; Tang, L.; Liu, M. The Hedyotis diffusa Willd. (Rubiaceae): A review on phytochemistry, pharmacology, quality control and pharmacokinetics. Molecules 2016, 21, 710. [Google Scholar] [CrossRef]
- Shu, M.H.; Appleton, D.; Zandi, K.; AbuBakar, S. Anti-inflammatory, gastroprotective and antiulcerogenic effects of red algae Gracilaria changii (Gracilariales, Rhodophyta) extract. BMC Complement. Altern. Med. 2013, 13, 61. [Google Scholar] [CrossRef]
- Urbatzka, R.; Freitas, S.; Palmeira, A.; Almeida, T.; Moreira, J.; Azevedo, C.; Afonso, C.; Correia-da-Silva, M.; Sousa, E.; Pinto, M.; et al. Lipid reducing activity and toxicity profiles of a library of polyphenol derivatives. Eur. J. Med. Chem. 2018, 151, 272–284. [Google Scholar] [CrossRef] [PubMed]
- Noinart, J.; Buttachon, S.; Dethoup, T.; Gales, L.; Pereira, J.A.; Urbatzka, R.; Freitas, S.; Lee, M.; Silva, A.M.S.; Pinto, M.M.M.; et al. A new ergosterol analog, a new bis-anthraquinone and anti-obesity activity of anthraquinones from the marine sponge-associated fungus Talaromyces stipitatus KUFA 0207. Mar. Drugs 2017, 16, 15. [Google Scholar] [CrossRef] [PubMed]
- Yao, L.; Gerde, J.A.; Lee, S.L.; Wang, T.; Harrata, K.A. Microalgae lipid characterization. J. Agric. Food Chem. 2015, 63, 1773–1787. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.; Park, J.; Kim, H.-L.; Kang, J.; Jeong, M.-Y.; Youn, D.-H.; Jung, Y.; Kim, Y.-I.; Kim, H.-J.; Ahn, K.S.; et al. Chrysophanic acid suppresses adipogenesis and induces thermogenesis by activating AMP-activated protein kinase alpha in vivo and in vitro. Front. Pharmacol. 2016, 7, 476. [Google Scholar] [CrossRef] [PubMed]
- Klingelhutz, A.J.; Gourronc, F.A.; Chaly, A.; Wadkins, D.A.; Burand, A.J.; Markan, K.R.; Idiga, S.O.; Wu, M.; Potthoff, M.J.; Ankrum, J.A. Scaffold-free generation of uniform adipose spheroids for metabolism research and drug discovery. Sci. Rep. 2018, 8, 523. [Google Scholar] [CrossRef] [Green Version]
- Turner, P.A.; Harris, L.M.; Purser, C.A.; Baker, R.C.; Janorkar, A.V. A surface-tethered spheroid model for functional evaluation of 3T3-L1 adipocytes. Biotechnol. Bioeng. 2014, 111, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Hihi, A.K.; Michalik, L.; Wahli, W. PPARs: Transcriptional effectors of fatty acids and their derivatives. Cell. Mol. Life Sci. 2002, 59, 790–798. [Google Scholar] [CrossRef]
- Fraher, D.; Sanigorski, A.; Mellett, N.A.; Meikle, P.J.; Sinclair, A.J.; Gibert, Y. Zebrafish embryonic lipidomic analysis reveals that the yolk cell is metabolically active in processing lipid. Cell Rep. 2016, 14, 1317–1329. [Google Scholar] [CrossRef]
- Chiarelli, F.; Di Marzio, D. Peroxisome proliferator-activated receptor-γ agonists and diabetes: Current evidence and future perspectives. Vasc. Health Risk Manag. 2008, 4, 297–304. [Google Scholar] [CrossRef]
- Chen, K.; Roca, M. In vitro bioavailability of chlorophyll pigments from edible seaweeds. J. Funct. Foods 2018, 41, 25–33. [Google Scholar] [CrossRef]
- Vieira, I.; Chen, K.; Ríos, J.J.; Benito, I.; Pérez-Gálvez, A.; Roca, M. First-Pass Metabolism of Chlorophylls in Mice. Mol. Nutr. Food Res. 2018, 17, e1800562. [Google Scholar] [CrossRef] [PubMed]
- Henriques, M.; Silva, A.; Rocha, J. Extraction and quantification of pigments from a marine microalga: A simple and reproducible method. Appl. Microbiol. 2007, 2, 586–893. [Google Scholar]
- Carpenter, A.E.; Jones, T.R.; Lamprecht, M.R.; Clarke, C.; Kang, I.H.; Friman, O.; Guertin, D.A.; Chang, J.H.; Lindquist, R.A.; Moffat, J.; et al. CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006, 7, R100. [Google Scholar] [CrossRef] [PubMed]
- Urbatzka, R.; Galante-Oliveira, S.; Rocha, E.; Castro, L.F.; Cunha, I. Normalization strategies for gene expression studies by real-time PCR in a marine fish species, Scophthalmus maximus. Mar. Genom. 2013, 10, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Freitas, S.; Martins, R.; Costa, M.; Leão, P.N.; Vitorino, R.; Vasconcelos, V.; Urbatzka, R. Hierridin B isolated from a marine cyanobacterium alters VDAC1, mitochondrial activity, and cell cycle genes on HT-29 colon adenocarcinoma cells. Mar. Drugs 2016, 14, 158. [Google Scholar] [CrossRef] [PubMed]





| Position | δH (J in Hz) | δC | Position | δH (J in Hz) | δC |
|---|---|---|---|---|---|
| 1 | 141.3 | 15 | 102.0/100.4 2 | ||
| 2 | 131.5 | 16 | 166.3 | ||
| 2 1 | 3.44 s | 11.5 | 17 | 4.07 dd (7.4) | 53.1 |
| 3 | 133.4 | 17 1 | 2.56 dd (7.7) 1.83 dd (9.1) | 30.7 | |
| 3 1 | 8.03 dd (11.5, 17.8) | 128.3 | 17 2 | 2.46 m 2.17 m | 31.7 |
| 3 2 | 6.35 d (18.7) 6.19 d (11.4) | 122.1 | 173 | 173.4 | |
| 4 | 135.9 | 18 | 4.46 m (7.8) | 49.7 | |
| 5 | 9.56 s | 99.1 | 18 1 | 1.59 m | 21.7 |
| 6 | 155.7 | 19 | 171 | ||
| 7 | 136.6 | 20 | 8.71 s | 93.3 | |
| 7 1 | 3.28 s | 10.8 | F1 | 4.5 m 4.44 m (6.8) | 60.8 |
| 8 | 145.6 | F2 | 5.14 t (6.5) | 117.1 | |
| 8 1 | 3.77 m 3.64 m | 19.0 | F3 | ||
| 8 2 | 1.72 t (7.7) | 17.0 | F3 1 | 1.61 s | 15.8 |
| 9 | 149.9 | F4 | 1.89 m (7.7) | 39.9 | |
| 10 | 9.77 s | 103.4 | F5 | 1.29 m (7.4) | 22-24 1 |
| 11 | 128.8 | F6 | 1.01 m | 36.6 | |
| 12 | 131.5 | F7 | 1.33 m | 32.1 | |
| 12 1 | 3.90 s | 11.9 | F7 1 | 0.8 m | 19.2 |
| 13 | 133.2 | F8 | 2.35 t (7.5) | 32.6 | |
| 13 1 | 1 | F9 | 1.65 m (7.3) | 24.3 | |
| 13 2-OH | 6.11 s | 110.4/102.0 2 | F10 | 1.11 m | 38.9 |
| 133 | 170.8 | F11 | 1.51 m | 27.4 | |
| 134-OCH3 | 3.76 s | 53.4 | F11 1 | 0.85 m (6.6) | 22.1 |
| 14 | 140.2 | F12 | 0.85 m (6.6) | 22.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Freitas, S.; Silva, N.G.; Sousa, M.L.; Ribeiro, T.; Rosa, F.; Leão, P.N.; Vasconcelos, V.; Reis, M.A.; Urbatzka, R. Chlorophyll Derivatives from Marine Cyanobacteria with Lipid-Reducing Activities. Mar. Drugs 2019, 17, 229. https://doi.org/10.3390/md17040229
Freitas S, Silva NG, Sousa ML, Ribeiro T, Rosa F, Leão PN, Vasconcelos V, Reis MA, Urbatzka R. Chlorophyll Derivatives from Marine Cyanobacteria with Lipid-Reducing Activities. Marine Drugs. 2019; 17(4):229. https://doi.org/10.3390/md17040229
Chicago/Turabian StyleFreitas, Sara, Natália Gonçalves Silva, Maria Lígia Sousa, Tiago Ribeiro, Filipa Rosa, Pedro N. Leão, Vitor Vasconcelos, Mariana Alves Reis, and Ralph Urbatzka. 2019. "Chlorophyll Derivatives from Marine Cyanobacteria with Lipid-Reducing Activities" Marine Drugs 17, no. 4: 229. https://doi.org/10.3390/md17040229





