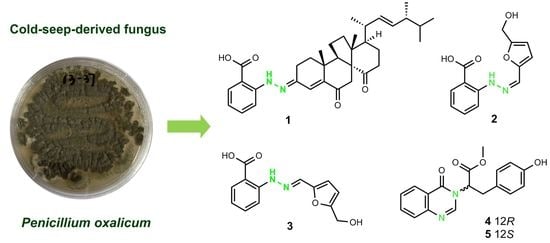
Phenylhydrazone and Quinazoline Derivatives from the Cold-Seep-Derived Fungus Penicillium oxalicum
Abstract
:1. Introduction
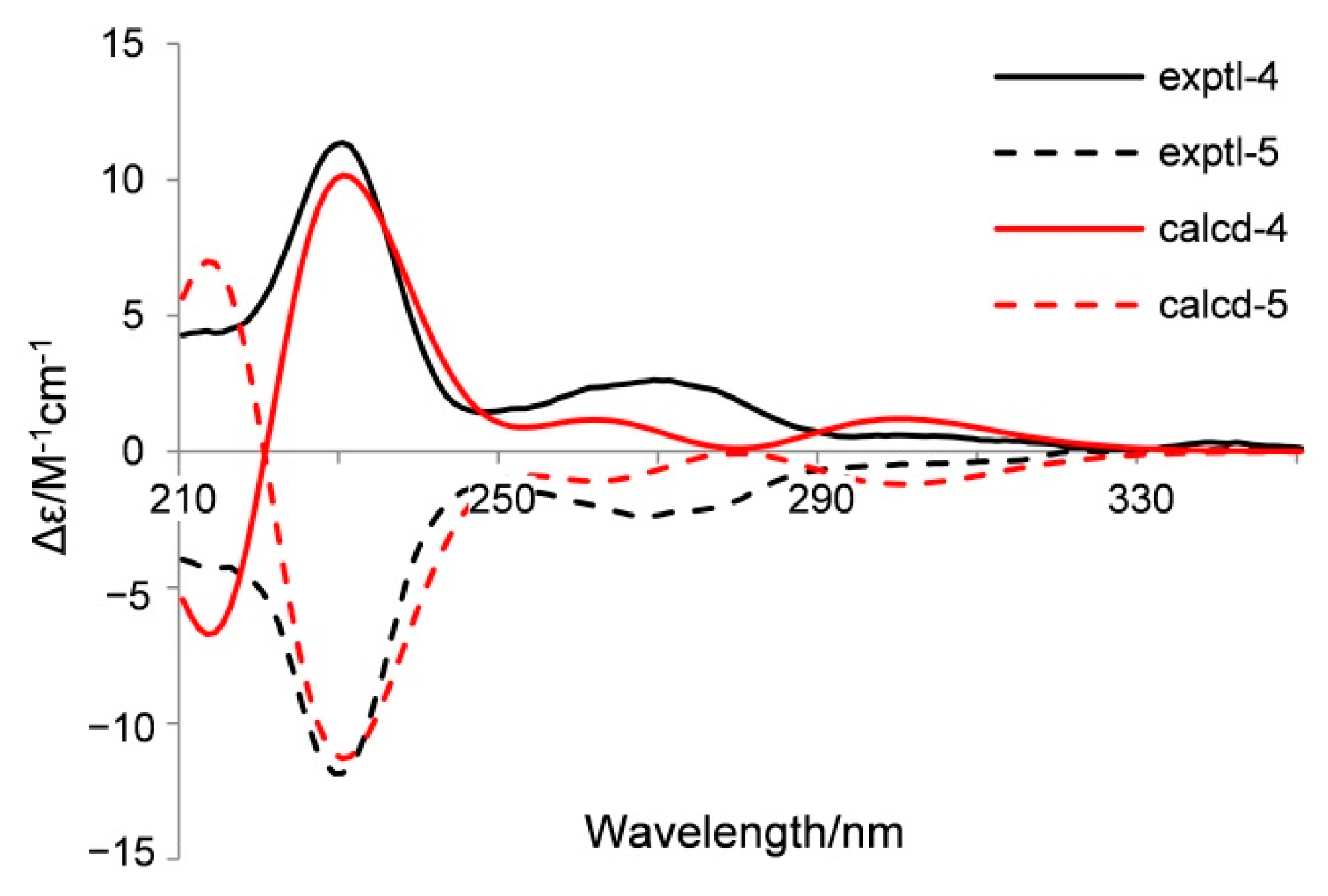
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material and Fermentation
3.3. Extraction and Isolation
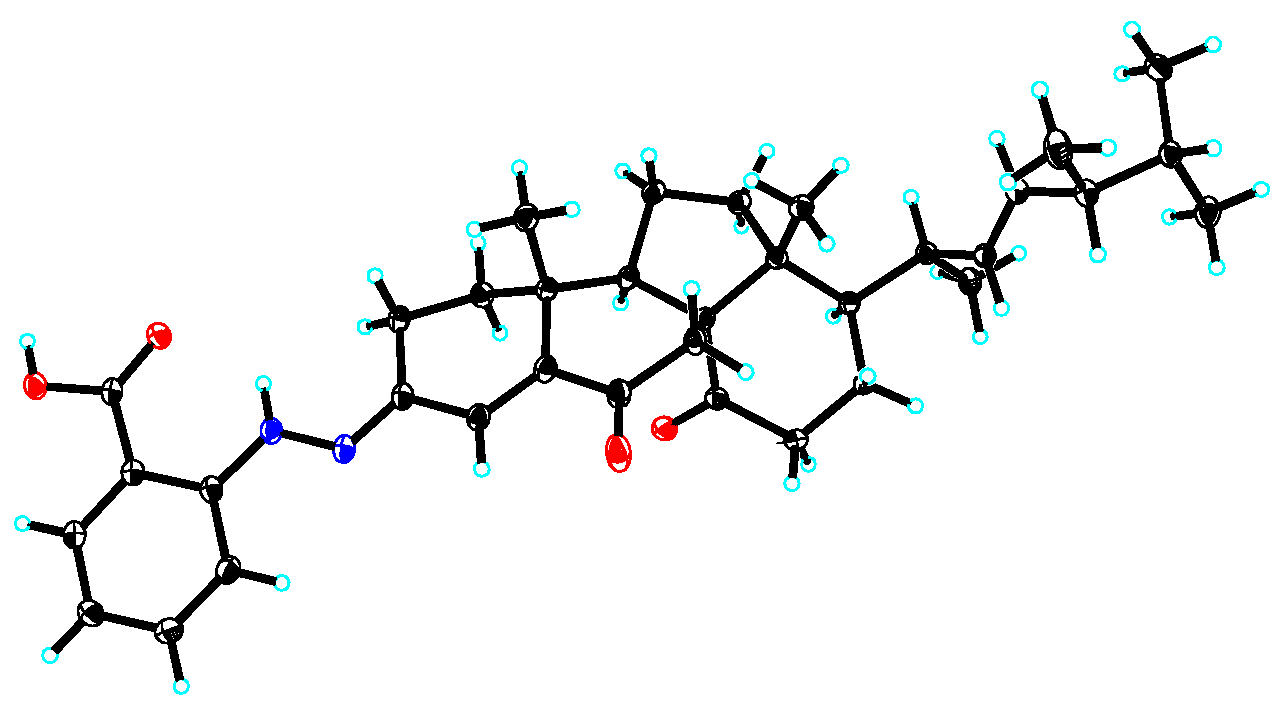
3.4. X-ray Crystallographic Analysis
3.5. Crystal Data for Penoxahydrazone A (1)
3.6. Computational Details
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Caneschi, W.L.; Sanchez, A.B.; Felestrino, É.B.; de Carvalho Lemes, C.G.; Cordeiro, I.F.; Fonseca, N.P.; Villa, M.M.; Vieira, I.T.; Moraes, L.Â.G.; de Almeida Barbosa Assis, R.; et al. Serratia liquefaciens FG3 isolated from a metallophyte plant sheds light on the evolution and mechanisms of adaptive traits in extreme environments. Sci. Rep. 2019, 9, 18006. [Google Scholar] [CrossRef] [PubMed]
- Bérdy, J. Bioactive microbial metabolites. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, T.; Zhou, Y.; Qin, J.J.; Yang, L.L.; Zhang, W.D.; Shen, Y.H. Chemical constituents from wetland soil fungus Penicillium oxalicum GY1. Fitoterapia 2020, 142, 104530. [Google Scholar] [CrossRef] [PubMed]
- Bian, X.Q.; Bai, J.; Hu, X.L.; Wu, X.; Xue, C.M.; Han, A.H.; Su, G.Y.; Hua, H.M.; Pei, Y.H. Penioxalicin, a novel 3-nor-2,3-seco-labdane type diterpene from the fungus Penicillium oxalicum TW01-1. Tetrahedron Lett. 2015, 56, 5013–5016. [Google Scholar] [CrossRef]
- Liu, B.; Wang, H.F.; Zhang, L.H.; Liu, F.; He, F.J.; Bai, F.J.; Hua, H.M.; Chen, G.; Pei, Y.H. Isolation of a new compound from Penicillium oxalicum. Chem. Nat. Compd. 2016, 52, 821–823. [Google Scholar] [CrossRef]
- Hu, X.L.; Bian, X.Q.; Wu, X.; Li, J.Y.; Hua, H.M.; Pei, Y.H.; Han, A.H.; Bai, J. Penioxalamine A, a novel prenylated spiro-oxindole alkaloid from Penicillium oxalicum TW01-1. Tetrahedron Lett. 2014, 55, 3864–3867. [Google Scholar] [CrossRef]
- Ren, Y.; Chao, L.H.; Sun, J.; Chen, X.N.; Yao, H.N.; Zhu, Z.X.; Dong, D.; Liu, T.; Tu, P.F.; Li, J. Two new polyketides from the fungus Penicillium oxalicum MHZ153. Nat. Prod. Res. 2018, 33, 347–353. [Google Scholar] [CrossRef]
- Seetharaman, P.; Gnanasekar, S.; Chandrasekaran, R.; Chandrakasan, G.; Syed, A.; Hodhod, M.S.; Ameen, F.; Sivaperumal, S. Isolation of limonoid compound (Hamisonine) from endophytic fungi Penicillium oxalicum LA-1 (KX622790). Environ. Sci. Pollut. Res. 2017, 24, 21272–21282. [Google Scholar] [CrossRef]
- Yuan, L.; Huang, W.Z.; Zhou, K.; Wang, Y.D.; Dong, W.; Du, G.; Gao, X.M.; Ma, Y.H.; Hu, Q.F. Butyrolactones derivatives from the fermentation products of a plant entophytic fungus Penicillium oxalicum. Nat. Prod. Res. 2015, 29, 1914–1919. [Google Scholar] [CrossRef]
- Li, Q.Q.; Dang, L.Z.; Zhang, Y.P.; Jiang, J.X.; Zhang, C.M.; Xiang, N.J.; Yang, H.Y.; Du, G.; Duan, Y.Q. Isocoumarins from the fermentation products of a plant entophytic fungus Penicillium oxalicum. J. Asian Nat. Prod. Res. 2015, 17, 876–881. [Google Scholar] [CrossRef]
- Yang, Z.; Huang, N.Y.; Xu, B.; Huang, W.F.; Xie, T.P.; Cheng, F.; Zou, K. Cytotoxic 1,3-thiazole and 1,2,4-thiadiazole alkaloids from Penicillium oxalicum: Structural elucidation and total synthesis. Molecules 2016, 21, 232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, S.; Guo, K.B.; Wang, X.Y.; Chen, H.; Min, J.B.; Qi, S.H.; Zhao, W.; Li, W.R. Toxicity study of oxalicumone A, derived from a marine-derived fungus Penicillium oxalicum, in cultured renal epithelial cells. Mol. Med. Rep. 2017, 15, 2611–2619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bao, J.; Zhang, X.Y.; Dong, J.J.; Xu, X.Y.; Nong, X.H.; Qi, S.H. Cyclopentane-condensed chromones from marine-derived fungus Penicillium oxalicum. Chem. Lett. 2014, 43, 837–839. [Google Scholar] [CrossRef]
- Bao, J.; Luo, J.F.; Qin, X.C.; Xu, X.Y.; Zhang, X.Y.; Tu, Z.C.; Qi, S.H. Dihydrothiophene-condensed chromones from a marine-derived fungus Penicillium oxalicum and their structure–bioactivity relationship. Bioorg. Med. Chem. Lett. 2014, 24, 2433–2436. [Google Scholar] [CrossRef]
- Sun, Y.L.; Bao, J.; Liu, K.S.; Zhang, X.Y.; He, F.; Wang, Y.F.; Nong, X.H.; Qi, S.H. Cytotoxic dihydrothiophene-condensed chromones from the marine-derived fungus Penicillium oxalicum. Planta Med. 2013, 79, 1474–1479. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Li, X.M.; Zhang, P.; Wang, B.G. A new phenolic enamide and a new meroterpenoid from marine alga-derived endophytic fungus Penicillium oxalicum EN-290. J. Asian Nat. Prod. Res. 2015, 17, 1204–1212. [Google Scholar] [CrossRef]
- Zhang, P.; Li, X.M.; Liu, H.; Li, X.; Wang, B.G. Two new alkaloids from Penicillium oxalicum EN-201, an endophytic fungus derived from the marine mangrove plant Rhizophora stylosa. Phytochem. Lett. 2015, 2015. 13, 160–164. [Google Scholar] [CrossRef]
- Wang, P.L.; Lib, D.Y.; Xie, L.R.; Wu, X.; Li, Z.L. Novel decaturin alkaloids from the marine-derived fungus Penicillium oxalicum. Nat. Prod. Commun. 2013, 8, 1397–1398. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Bi, Y.X.; Li, Y.P.; Li, X.X.; Liu, Q.Y.; Ying, M.G.; Zheng, Q.H.; Du, L.; Zhang, Q.Q. Secalonic acids H and I, two new secondary metabolites from the marine-derived fungus Penicillium oxalicum. Heterocycles 2017, 94, 1766–1774. [Google Scholar]
- Chen, L.; Lu, Z.H.; Liu, Q.Y.; Zheng, Q.H.; Du, L.; Zhang, Q.Q. Secalonic acids J–M, four new secondary metabolites from the marine-derived fungus Penicillium oxalicum. Heterocycles 2019, 98, 155–965. [Google Scholar]
- Liu, B.; Wang, H.F.; Zhang, L.H.; Liu, F.; He, F.J.; Bai, J.; Hua, H.M.; Chen, G.; Pei, Y.H. New compound with DNA Topo I inhibitory activity purified from Penicillium oxalicum HSY05. Nat. Prod. Res. 2015, 29, 2197–2202. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.L.; Li, D.Y.; Xie, L.R.; Wu, X.; Hua, H.M.; Li, Z.L. Two new compounds from a marine-derived fungus Penicillium oxalicum. Nat. Prod. Res. 2013, 28, 290–293. [Google Scholar] [CrossRef] [PubMed]
- Pimenta, E.F.; Vita-Marques, A.M.; Tininis, A.; Seleghim, M.H.R.; Sette, L.D.; Veloso, K.; Ferreira, A.G.; Williams, D.E.; Patrick, B.O.; Dalisay, D.S.; et al. Use of experimental design for the optimization of the production of new secondary metabolites by two Penicillium species. J. Nat. Prod. 2010, 73, 1821–1832. [Google Scholar] [CrossRef] [PubMed]
- Orsi, W.D.; Edgcomb, V.P.; Christman, G.D.; Biddle, J.F. Gene expression in the deep biosphere. Nature 2013, 499, 205–208. [Google Scholar] [CrossRef] [PubMed]
- Orsi, W.D.; Richards, T.A.; Santoro, A.E. Cellular maintenance processes that potentially underpin the survival of subseafloor fungi over geological timescales. Estuar. Coast. Shelf Sci. 2015, 164, A1–A9. [Google Scholar] [CrossRef]
- Amagata, T.; Tanaka, M.; Yamada, T.; Doi, M.; Minoura, K.; Ohishi, H.; Yamori, T.; Numata, A. Variation in cytostatic constituents of a sponge-derived Gymnascella dankaliensis by manipulating the carbon source. J. Nat. Prod. 2007, 70, 1731–1740. [Google Scholar] [CrossRef]
- Cheng, P.; Xu, K.; Chen, Y.C.; Wang, T.T.; Chen, Y.; Yang, C.L.; Ma, S.Y.; Liang, Y.; Ge, H.M.; Jiao, R.H. Cytotoxic aromatic polyketides from an insect derived Streptomyces sp. NA4286. Tetrahedron Lett. 2019, 60, 1706–1709. [Google Scholar] [CrossRef]
- Song, Y.P.; Shi, Z.Z.; Miao, F.P.; Fang, S.T.; Yin, X.L.; Ji, N.Y. Tricholumin A, a highly transformed ergosterol derivative from the alga-endophytic fungus Trichoderma asperellum. Org. Lett. 2018, 20, 6306–6309. [Google Scholar] [CrossRef]
- Blair, L.M.; Sperry, J. Natural products containing a nitrogen−nitrogen bond. J. Nat. Prod. 2013, 76, 794–812. [Google Scholar] [CrossRef]
- Han, M.; Liu, X.; Zhang, X.; Pang, Y.; Xu, P.; Guo, J.; Liu, Y.; Zhang, S.; Ji, S. 5-Hydroxymethyl-2-vinylfuran: A biomass-based solvent-free adhesive. Green Chem. 2017, 19, 722–728. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision C.01; Gaussian, Inc.: Wallingford, CT, USA, 2010. [Google Scholar]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Beyond DP4: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 2015, 80, 12526–12534. [Google Scholar] [CrossRef] [PubMed]
- Rao, K.R.; Raghunadh, A.; Mekala, R.; Meruva, S.B.; Pratap, T.V.; Krishna, T.; Kalita, D.; Laxminarayana, E.; Prasad, B.; Pal, M. Glyoxylic acid in the reaction of isatoic anhydride with amines: A rapid synthesis of 3-(un)substituted quinazolin-4(3h)-ones leading to rutaecarpine and evodiamine. Tetrahedron Lett. 2014, 55, 6004–6006. [Google Scholar] [CrossRef]
- Allegretta, G.; Weidel, E.; Empting, M.; Hartmann, R.W. Catechol-based substrates of chalcone synthase as a scaffold for novel inhibitors of PqsD. Eur. J. Med. Chem. 2015, 90, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.P.; Miao, F.P.; Fang, S.T.; Yin, X.L.; Ji, N.Y. Halogenated and nonhalogenated metabolites from the marine-alga-endophytic fungus Trichoderma asperellum cf44-2. Mar. Drugs 2018, 16, 266. [Google Scholar] [CrossRef] [Green Version]
- Miao, F.P.; Liang, X.R.; Yin, X.L.; Wang, G.; Ji, N.Y. Absolute configurations of unique harziane diterpenes from Trichoderma species. Org. Lett. 2012, 14, 3815–3817. [Google Scholar] [CrossRef]
- Pu, D.B.; Zhang, X.J.; Bi, D.W.; Gao, J.B.; Yang, Y.; Li, X.L.; Lin, J.; Li, X.N.; Zhang, R.H.; Xiao, W.L. Callicarpins, two classes of rearranged ent-clerodane diterpenoids from Callicarpa plants blocking NLRP3 inflammasome-induced pyroptosis. J. Nat. Prod. 2020, 83, 2191–2199. [Google Scholar] [CrossRef]
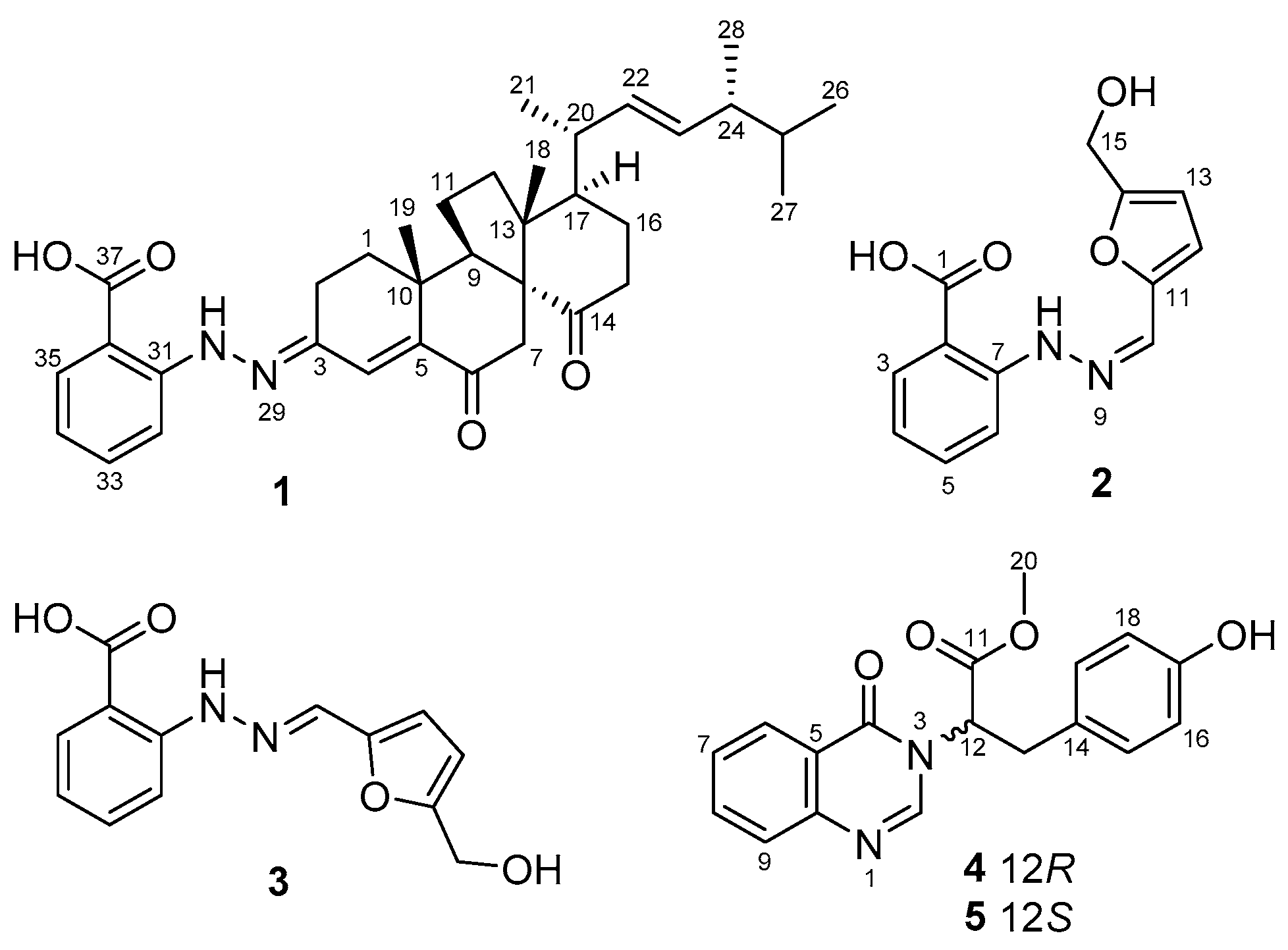

| Pos | δc, Type | δH (J in Hz) |
|---|---|---|
| 1a | 37.6, CH2 | 1.98, m |
| 1b | 1.90, m | |
| 2a | 20.9, CH2 | 2.60, m |
| 2b | 2.40, m | |
| 3 | 144.4, C | |
| 4 | 131.7, CH | 7.08, s |
| 5 | 141.6, C | |
| 6 | 199.2, C | |
| 7a | 41.3, CH2 | 2.61, d (16.1) |
| 7b | 2.53, d (16.1) | |
| 8 | 62.8, C | |
| 9 | 49.5, CH | 2.85, t (9.4) |
| 10 | 35.6, C | |
| 11a | 25.6, CH2 | 1.92, m |
| 11b | 1.81, m | |
| 12a | 39.2, CH2 | 1.75, m |
| 12b | 1.62, dt (12.5, 8.1) | |
| 13 | 53.8, C | |
| 14 | 215.9, C | |
| 15a | 38.2, CH2 | 2.56, m |
| 15b | 2.46, m | |
| 16a | 23.2, CH2 | 1.86, m |
| 16b | 1.74, m | |
| 17 | 50.8, CH | 1.39, br d (12.7) |
| 18 | 17.3, CH3 | 1.00, s |
| 19 | 24.5, CH3 | 1.16, s |
| 20 | 37.5, CH | 2.39, m |
| 21 | 23.6, CH3 | 1.07, d (7.0) |
| 22 | 132.8, CH | 5.25, dd (15.5, 4.6) |
| 23 | 135.0, CH | 5.28, dd (15.5, 5.0) |
| 24 | 43.4, CH | 1.87, m |
| 25 | 33.2, CH | 1.47, octet (6.8) |
| 26 | 19.8, CH3 | 0.82, d (6.8) |
| 27 | 20.2, CH3 | 0.84, d (6.8) |
| 28 | 17.8, CH3 | 0.92, d (6.8) |
| 30 | 11.07, s | |
| 31 | 147.3, C | |
| 32 | 114.1, CH | 7.66, d (8.5) |
| 33 | 135.8, CH | 7.45, dd (8.5, 7.1) |
| 34 | 119.1, CH | 6.84, dd (7.8, 7.1) |
| 35 | 131.7, CH | 7.98, d (7.8) |
| 36 | 109.4, C | |
| 37 | 172.4, C |
| Pos | 2 | 3 | ||
|---|---|---|---|---|
| δc, Type | δH (J in Hz) | δc, Type | δH (J in Hz) | |
| 1 | 169.6, C | 169.5, C | ||
| 2 | 111.0, C | 110.1, C | ||
| 3 | 131.3, CH | 7.89, dd (7.9, 1.3) | 131.2, CH | 7.84, dd (7.9, 1.4) |
| 4 | 118.3, CH | 6.84, br dd (7.9, 7.1) | 117.6, CH | 6.79, dd (7.9, 7.1) |
| 5 | 134.4, CH | 7.48, br dd (8.4, 7.1) | 134.4, CH | 7.48, br dd (8.4, 7.1) |
| 6 | 112.9, CH | 7.68, br d (8.4) | 113.1, CH | 7.59, br d (8.4) |
| 7 | 147.1, C | 146.9, C | ||
| 8 | 12.33, br s | 11.23, br s | ||
| 10 | 125.4, CH | 7.27, s | 131.4, CH | 8.03, s |
| 11 | 146.8, C | 149.4, C | ||
| 12 | 113.7, CH | 7.01, d (3.3) | 112.0, CH | 6.65, d (3.3) |
| 13 | 108.9, CH | 6.54, d (3.3) | 109.2, CH | 6.40, d (3.3) |
| 14 | 157.2, C | 156.7, C | ||
| 15 | 56.0, CH2 | 4.57, s | 55.8, CH2 | 4.44, s |
| Pos | δc, Type | δH (J in Hz) |
|---|---|---|
| 2 | 147.2, CH | 8.01, s |
| 4 | 159.9, C | |
| 5 | 121.1, C | |
| 6 | 126.1, CH | 8.12, dd (7.9, 1.2) |
| 7 | 127.4, CH | 7.55, dd (7.9, 7.1) |
| 8 | 134.8, CH | 7.83, ddd (8.1, 7.1, 1.2) |
| 9 | 127.2, CH | 7.62, d (8.1) |
| 10 | 147.4, C | |
| 11 | 169.3, C | |
| 12 | 60.4, CH | 5.41, dd (11.1, 4.9) |
| 13a | 33.3, CH2 | 3.41, dd (14.3, 4.9) |
| 13b | 3.32, dd (14.3, 11.1) | |
| 14 | 126.1, C | |
| 15 | 129.8, CH | 6.87, d (8.4) |
| 16 | 115.3, CH | 6.54, d (8.4) |
| 17 | 156.2, C | |
| 18 | 115.3, CH | 6.54, d (8.4) |
| 19 | 129.8, CH | 6.87, d (8.4) |
| 20 | 52.6, CH3 | 3.69, s |
| Compounds | IC50 (μg/mL) | Inhibitory Zone Diameter (mm) at 20 μg/disk | LC50 (μg/mL) | |||||
|---|---|---|---|---|---|---|---|---|
| C. marina | H. akashiwo | P. donghaiense | V. anguillarum | V. harveyi | V. parahaemolyticus | V. splendidus | A. salina | |
| 1 | 1.2 | 3.7 | 0.68 | 0 | 11 | 12 | 0 | 58 |
| 2/3 | 17 | >100 | 5.4 | 0 | 0 | 7.0 | 9.7 | >100 |
| 4 | 2.8 | 8.1 | 0.57 | 7.0 | 13 | 11 | 7.3 | >100 |
| 5 | 9.1 | 9.0 | 1.2 | 0 | 13 | 11 | 7.0 | >100 |
| dankasterone A | 1.9 | 4.6 | 1.0 | 7.0 | 12 | 12 | 0 | 43 |
| K2Cr2O7 | 0.60 | 2.4 | 1.2 | 18 | ||||
| chloramphenicol | 18 | 29 | 28 | 23 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.-P.; Fang, S.-T.; Shi, Z.-Z.; Wang, B.-G.; Li, X.-N.; Ji, N.-Y. Phenylhydrazone and Quinazoline Derivatives from the Cold-Seep-Derived Fungus Penicillium oxalicum. Mar. Drugs 2021, 19, 9. https://doi.org/10.3390/md19010009
Liu Y-P, Fang S-T, Shi Z-Z, Wang B-G, Li X-N, Ji N-Y. Phenylhydrazone and Quinazoline Derivatives from the Cold-Seep-Derived Fungus Penicillium oxalicum. Marine Drugs. 2021; 19(1):9. https://doi.org/10.3390/md19010009
Chicago/Turabian StyleLiu, Ya-Ping, Sheng-Tao Fang, Zhen-Zhen Shi, Bin-Gui Wang, Xiao-Nian Li, and Nai-Yun Ji. 2021. "Phenylhydrazone and Quinazoline Derivatives from the Cold-Seep-Derived Fungus Penicillium oxalicum" Marine Drugs 19, no. 1: 9. https://doi.org/10.3390/md19010009