SpCrus2 Glycine-Rich Region Contributes Largely to the Antiviral Activity of the Whole-Protein Molecule by Interacting with VP26, a WSSV Structural Protein
Abstract
:1. Introduction
2. Results
2.1. Bioinformatic Analysis of SpCrus2 GRR
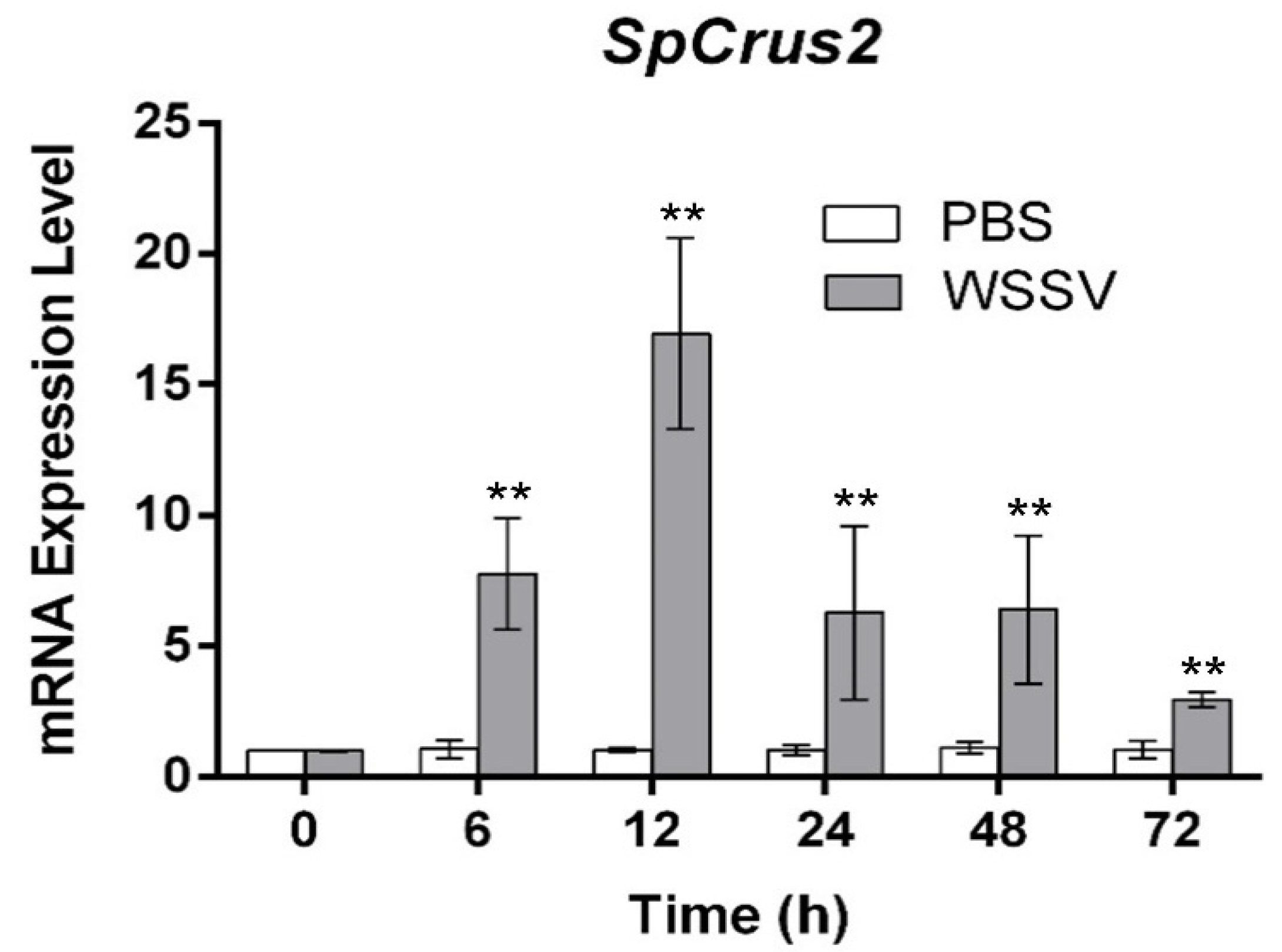
2.2. SpCrus2 Was Upregulated after WSSV Challenge
2.3. SpCrus2 Knockdown Facilitated WSSV Proliferation
2.4. WSSV Structure Proteins Were Recombinantly Expressed in a Soluble Form
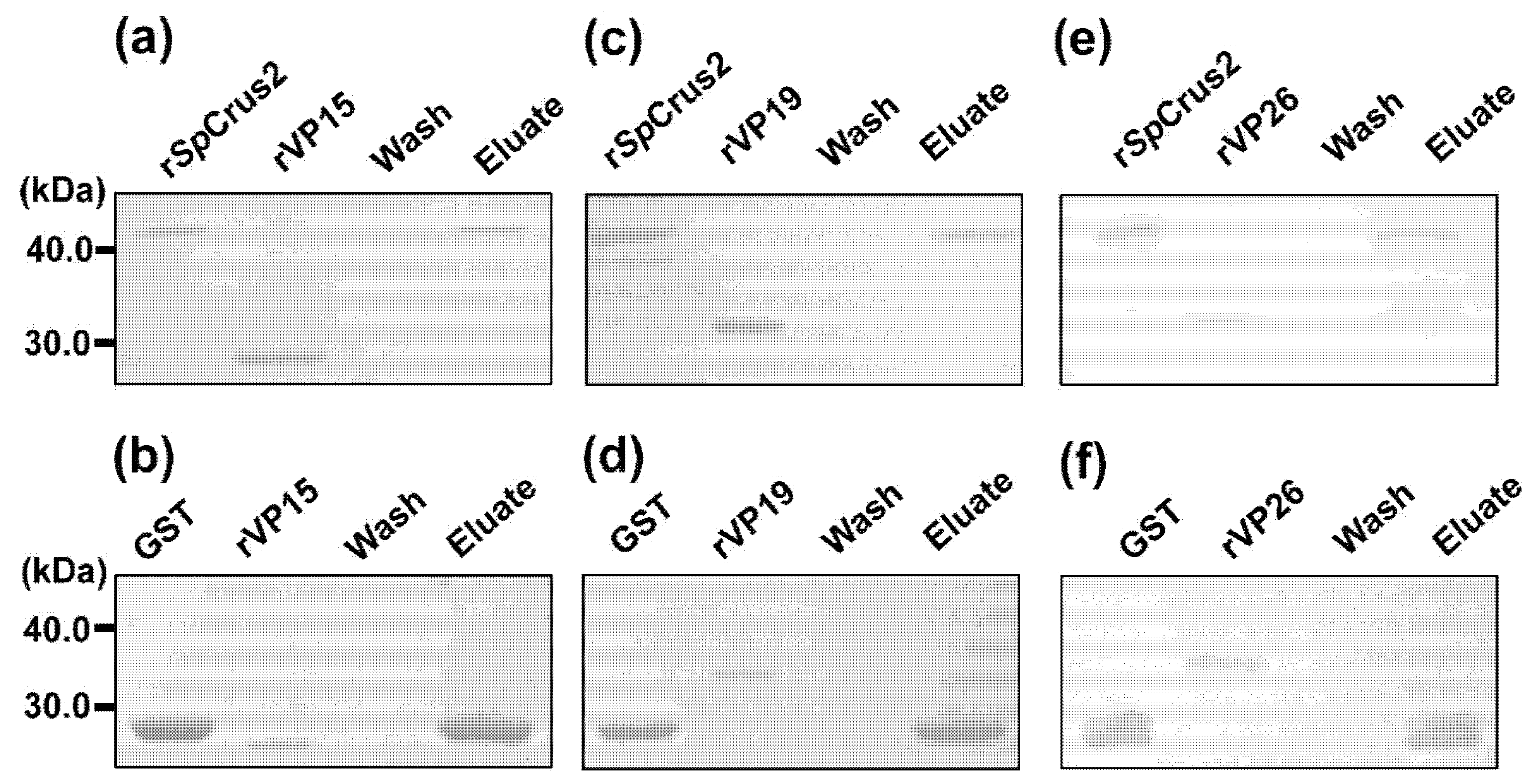
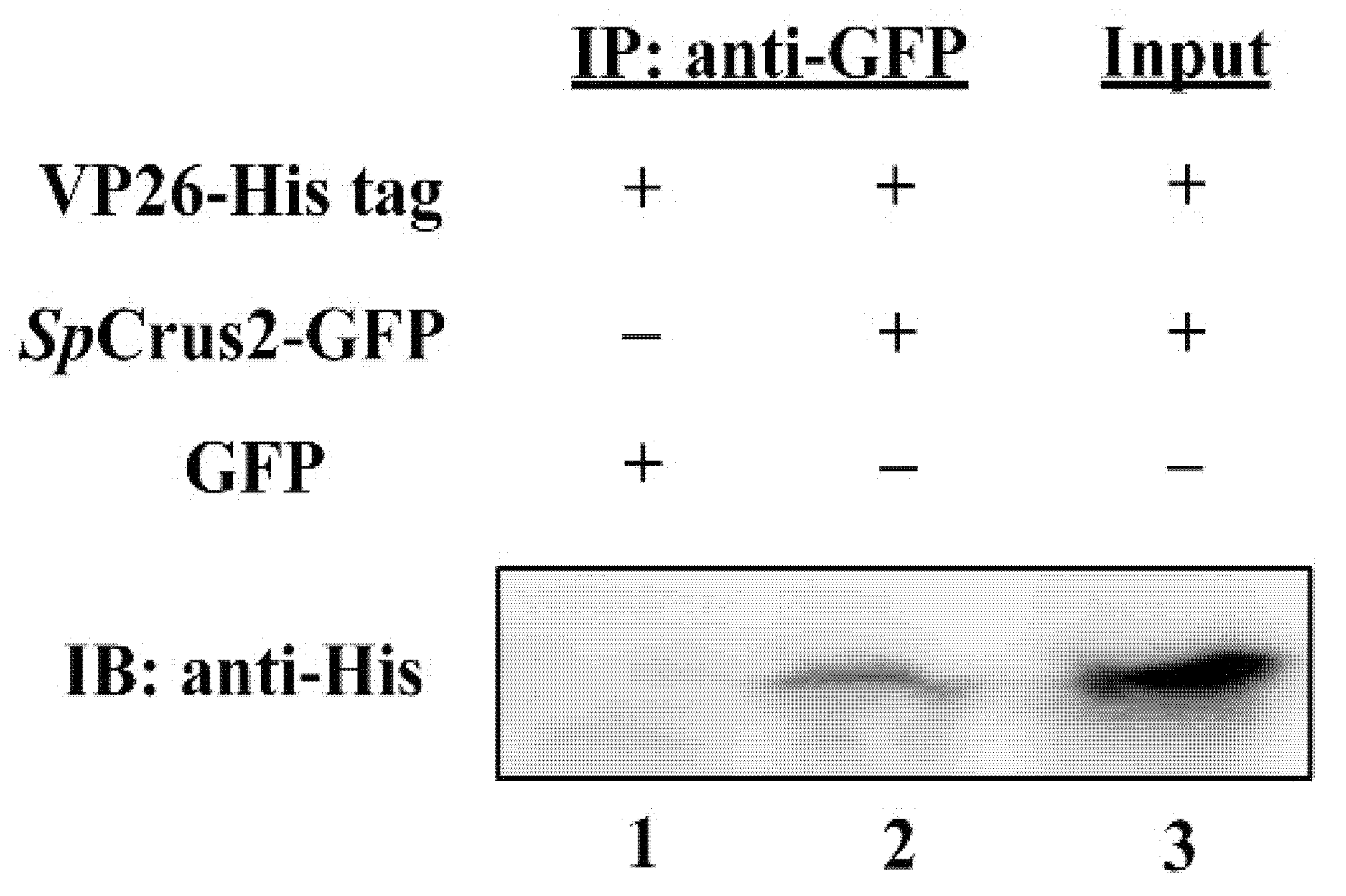
2.5. SpCrus2 Interacted Specifically with the WSSV Structural Protein VP26
2.6. SpCrus2 GRR Showed Stronger Binding Affinity to VP26 Than Its CRR
2.7. SpCrus2 GRR Exhibited Stronger Inhibitory Activity on WSSV Proliferation Than Its CRR
3. Discussion
4. Materials and Methods
4.1. Reagents and Chemicals
4.2. Mud Crab Challenge and Tissue Collection
4.3. Total RNA Isolation and cDNA Synthesis
4.4. Bioinformatic Analysis
4.5. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
4.6. RNA Interference (RNAi) of SpCrus2
4.7. Recombinant Expression and Purification of Viral Proteins, SpCrus2, GRR, and CRR
4.8. Profiles of WSSV Proliferation Affected by RNAi of SpCrus2 or by Protein Pre-Incubation
4.9. GST Pull-Down Assay
4.10. Plasmid Constructions, Cell Culture, and Co-IP Assay
4.11. ELISA
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Hancock, R.E.; Diamond, G. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 2000, 8, 402–410. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef]
- Hancock, R.E.; Sahl, H.G. Antimicrobial and host-defense peptides as new anti-infective therapeutic strategies. Nat. Biotechnol. 2006, 24, 1551–1557. [Google Scholar] [CrossRef] [PubMed]
- Tassanakajon, A.; Amparyup, P.; Somboonwiwat, K.; Supungul, P. Cationic antimicrobial peptides in penaeid shrimp. Mar. Biotechnol. 2010, 12, 487–505. [Google Scholar] [CrossRef] [PubMed]
- Smith, V.J.; Fernandes, J.M.; Kemp, G.D.; Hauton, C. Crustins: Enigmatic WAP domain-containing antibacterial proteins from crustaceans. Dev. Comp. Immunol. 2008, 32, 758–772. [Google Scholar] [CrossRef] [Green Version]
- Hou, Z.G.; Wang, Y.; Hui, K.; Fang, W.H.; Zhao, S.; Zhang, J.X.; Ma, H.; Li, X.C. A novel anti-lipopolysaccharide factor SpALF6 in mud crab Scylla paramamosain exhibiting different antimicrobial activity from its single amino acid mutant. Dev. Comp. Immunol. 2017, 72, 44–56. [Google Scholar] [CrossRef]
- Zhou, J.; Zhao, S.; Fang, W.H.; Zhou, J.F.; Zhang, J.X.; Ma, H.; Lan, J.F.; Li, X.C. Newly identified invertebrate-type lysozyme (Splys-i) in mud crab (Scylla paramamosain) exhibiting muramidase-deficient antimicrobial activity. Dev. Comp. Immunol. 2017, 74, 154–166. [Google Scholar] [CrossRef] [PubMed]
- Tassanakajon, A.; Somboonwiwat, K.; Amparyup, P. Sequence diversity and evolution of antimicrobial peptides in invertebrates. Dev. Comp. Immunol. 2015, 48, 324–341. [Google Scholar] [CrossRef]
- Li, H.; Yin, B.; Wang, S.; Fu, Q.; Xiao, B.; Lǚ, K.; He, J.; Li, C. RNAi screening identifies a new Toll from shrimp Litopenaeus vannamei that restricts WSSV infection through activating Dorsal to induce antimicrobial peptides. PLoS Pathog. 2018, 14, e1007109. [Google Scholar] [CrossRef] [Green Version]
- Leu, J.H.; Yang, F.; Zhang, X.; Xu, X.; Kou, G.H.; Lo, C.F. Whispovirus. Curr. Top. Microbiol. Immunol. 2009, 328, 197–227. [Google Scholar]
- Tan, Y.W.; Shi, Z.L. Proteomic analyses of the shrimp white spot syndrome virus. Virol. Sin. 2008, 3, 157–166. [Google Scholar] [CrossRef]
- Tsai, J.M.; Wang, H.C.; Leu, J.H.; Wang, A.H.; Zhuang, Y.; Walker, P.J.; Kou, G.H.; Lo, C.F. Identification of the nucleocapsid, tegument, and envelope proteins of the shrimp white spot syndrome virus virion. J. Virol. 2006, 80, 3021–3029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.; Hong, Y.; Qiu, H.; Yang, F.; Li, F. VP19 is important for the envelope coating of white spot syndrome virus. Virus Res. 2019, 270, 197666. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Xu, L.; Yang, F. VP26 of white spot syndrome virus functions as a linker protein between the envelope and nucleocapsid of virions by binding with VP51. J. Virol. 2008, 82, 12598–12601. [Google Scholar] [CrossRef] [Green Version]
- Relf, J.M.; Chisholm, J.R.; Kemp, G.D.; Smith, V.J. Purification and characterization of a cysteine-rich 11.5-kDa antibacterial protein from the granular haemocytes of the shore crab, Carcinus maenas. Eur. J. Biochem. 1999, 264, 350–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suthianthong, P.; Donpudsa, S.; Supungul, P.; Tassanakajon, A.; Rimphanitchayakit, V. The N-terminal glycine-rich and cysteine-rich regions are essential for antimicrobial activity of crustinPm1 from the black tiger shrimp Penaeus monodon. Fish Shellfish Immunol. 2012, 33, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, J.X.; Wang, Y.; Fang, W.H.; Wang, Y.; Zhou, J.F.; Zhao, S.; Li, X.C. Newly identified type II crustin (SpCrus2) in Scylla paramamosain contains a distinct cysteine distribution pattern exhibiting broad antimicrobial activity. Dev. Comp. Immunol. 2018, 84, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Ma, C.; Zhu, P.; Yang, Y.; Lei, A.; Chen, X.; Liang, W.; Chen, M.; Xiong, J.; Li, C. A new crustin is involved in the innate immune response of shrimp Litopenaeus vannamei. Fish Shellfish Immunol. 2019, 94, 398–406. [Google Scholar] [CrossRef]
- Li, S.; Lv, X.; Yu, Y.; Zhang, X.; Li, F. Molecular and Functional Diversity of Crustin-Like Genes in the Shrimp Litopenaeus vannamei. Mar. Drugs 2020, 18, 361. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Huang, X.; Zhang, Z.; Zhang, R.; Cao, X.; Zhang, C.; Wang, K.; Ren, Q. Molecular cloning and expression analysis of two type II crustin genes in the oriental river prawn, Macrobrachium nipponense. Fish Shellfish Immunol. 2020, 98, 446–456. [Google Scholar] [CrossRef]
- Chen, F.; Wang, K. Characterization of the innate immunity in the mud crab Scylla paramamosain. Fish Shellfish Immunol. 2019, 93, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Imjongjirak, C.; Amparyup, P.; Tassanakajon, A.; Sittipraneed, S. Molecular cloning and characterization of crustin from mud crab Scylla paramamosain. Mol. Biol. Rep. 2009, 36, 841–850. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.W.; Wang, H.; Fang, W.H.; Ma, H.; Zhang, F.; Wang, Y.; Li, X.C. SpCrus3 and SpCrus4 share high similarity in mud crab (Scylla paramamosain) exhibiting different antibacterial activities. Dev. Comp. Immunol. 2018, 82, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, C.; Wang, H.; Ma, H.; Huang, Y.Q.; Lu, J.X.; Li, X.C.; Zhang, X.W. Involvement of a newly identified atypical type II crustin (SpCrus5) in the antibacterial immunity of mud crab Scylla paramamosain. Fish Shellfish Immunol. 2018, 75, 346–356. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.Q.; Wang, Y.; Ma, H.Y.; Shen, X.L.; Wang, K.; Du, J.; Yu, X.D.; Fang, W.H.; Li, X.C. A new crustin homologue (SpCrus6) involved in the antimicrobial and antiviral innate immunity in mud crab, Scylla paramamosain. Fish Shellfish Immunol. 2019, 84, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Vargas-Albores, F.; Martínez-Porchas, M. Crustins are distinctive members of the WAP-containing protein superfamily: An improved classification approach. Dev. Comp. Immunol. 2017, 76, 9–17. [Google Scholar] [CrossRef]
- Sachetto-Martins, G.; Franco, L.O.; de Oliveira, D.E. Plant glycine-rich proteins: A family or just proteins with a common motif? Biochim. Biophys. Acta 2000, 1492, 1–14. [Google Scholar] [CrossRef]
- Mangeon, A.; Junqueira, R.M.; Sachetto-Martins, G. Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal. Behav. 2010, 5, 99–104. [Google Scholar] [CrossRef] [Green Version]
- van der Weerden, N.L.; Bleackley, M.R.; Anderson, M.A. Properties and mechanisms of action of naturally occurring antifungal peptides. Cell. Mol. Life Sci. 2013, 70, 3545–3570. [Google Scholar] [CrossRef]
- Lin, F.Y.; Gao, Y.; Wang, H.; Zhang, Q.X.; Zeng, C.L.; Liu, H.P. Identification of an anti-lipopolysacchride factor possessing both antiviral and antibacterial activity from the red claw crayfish Cherax quadricarinatus. Fish Shellfish Immunol. 2016, 57, 213–221. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, S.; Zhang, B.; Ma, H.Y.; Fang, W.H.; Sheng, W.Q.; Yang, L.G.; Li, X.C. A novel ML domain-containing protein (SpMD2) functions as a potential LPS receptor involved in anti-Vibrio immune response. Dev. Comp. Immunol. 2020, 103, 103529. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.P.; Zhang, B.; Li, Z.M.; Yang, L.G.; Li, X.C.; Ma, H. A lysin motif-containing protein (SpLysMD3) functions as a PRR involved in the antibacterial responses of mud crab, Scylla paramamosain. Fish Shellfish Immunol. 2020, 97, 257–267. [Google Scholar] [CrossRef] [PubMed]
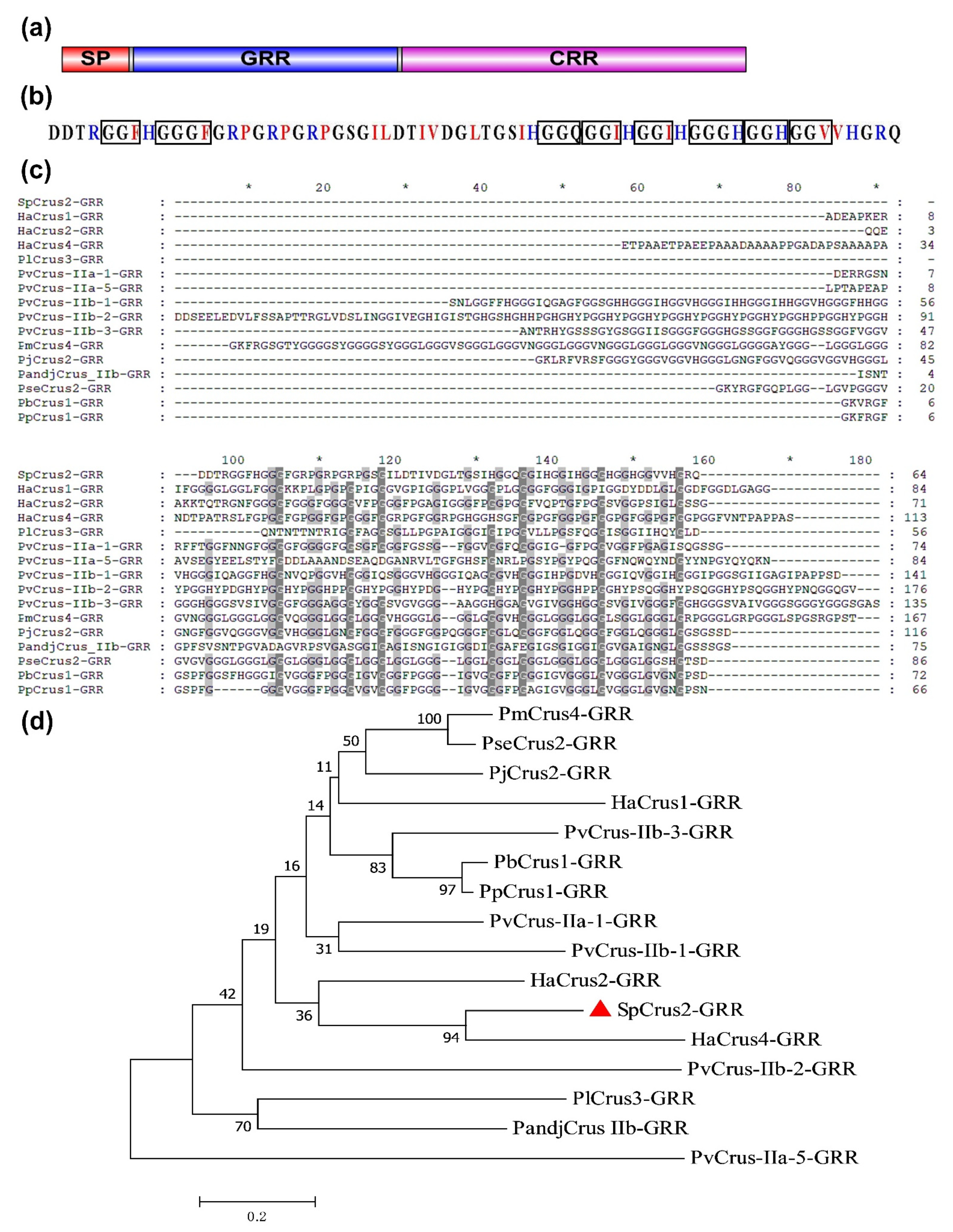




| Species | Gene Names | AAs Numbers | MWs (kDa) | pIs | Gly Contents | NCBI Accession Number of Genes |
|---|---|---|---|---|---|---|
| Scylla paramamosain | SpCrus2 | 64 | 6.1 | 9.44 | 40.6 | AUV47162 |
| Hyalella azteca | HaCrus1 | 84 | 7.3 | 4.19 | 47.6 | XP_018011791 |
| HaCrus2 | 71 | 6.3 | 9.99 | 45.1 | XP_018011805 | |
| HaCrus3 | 26 | 2.3 | 3.8 | 30.8 | XP_018011808 | |
| HaCrus4 | 113 | 10.1 | 4.69 | 31.9 | XP_018017888 | |
| Pacifastacus leniusculus | PlCrus3 | 56 | 5.3 | 6.74 | 32.1 | ABP88044 |
| Penaeus vannamei | PvCrus-IIa-1 | 74 | 6.6 | 9.51 | 50.0 | MT375570 |
| PvCrus-IIa-2 | 61 | 4.9 | 8.75 | 57.4 | AAL36890 | |
| PvCrus-IIa-3 | 63 | 5.5 | 8.59 | 42.9 | AFV77524 | |
| PvCrus-IIa-4 | 62 | 5.9 | 11.7 | 41.9 | MT375573 | |
| PvCrus-IIa-5 | 84 | 9.1 | 4.1 | 15.5 | MT375574 | |
| PvCrus-IIa-6 | 56 | 5.0 | 6.99 | 33.9 | MT375575 | |
| PvCrus-IIa-7 | 70 | 6.9 | 7.16 | 35.7 | MT375576 | |
| PvCrus-IIa-8 | 50 | 4.26 | 7.06 | 42.0 | MT375577 | |
| PvCrus-IIa-9 | 35 | 3.0 | 5.98 | 37.1 | MT375578 | |
| PvCrus-IIa-10 | 53 | 4.0 | 7.89 | 67.9 | MT375579 | |
| PvCrus-IIa-11 | 50 | 4.52 | 5.38 | 40.0 | MT375580 | |
| PvCrus-IIb-1 | 141 | 12.5 | 6.84 | 46.8 | MT375581 | |
| PvCrus-IIb-2 | 176 | 17.9 | 6.33 | 31.2 | MT375582 | |
| PvCrus-IIb-3 | 135 | 10.8 | 8.51 | 54.8 | MT375583 | |
| PvCrus1 | 49 | 4.1 | 10.84 | 55.1 | AAL36892 | |
| PvCrus2 | 61 | 5.0 | 8.75 | 55.7 | AAL36894 | |
| PvCrus3 | 67 | 5.4 | 8.75 | 58.2 | AAL36895 | |
| Penaeus monodon | PmCrus1 | 42 | 3.9 | 12.48 | 38.1 | ACQ66004 |
| PmCrus2 | 62 | 5.6 | 12.0 | 46.8 | XP_037772780 | |
| PmCrus4 | 167 | 13.0 | 10.53 | 63.5 | ACQ66005 | |
| PmCrus5 | 63 | 6.1 | 6.25 | 38.1 | ACP40176 | |
| PmCrus6 | 33 | 3.1 | 12.3 | 33.3 | ABW82154 | |
| PmCrus7 | 52 | 4.7 | 5.38 | 40.4 | ACL97375 | |
| PmCrus8 | 57 | 5.1 | 5.38 | 40.4 | ABV25094 | |
| Penaeus japonicus | PjCrus1 | 88 | 7.5 | 9.99 | 56.8 | BAD15062 |
| PjCrus2 | 116 | 9.8 | 9.99 | 57.8 | BAD15063 | |
| PjCrus3 | 108 | 9.2 | 9.99 | 57.4 | BAD15064 | |
| PjCrus4 | 104 | 8.8 | 9.99 | 57.7 | BAD15065 | |
| PjCrus5 | 80 | 6.8 | 9.99 | 56.2 | BAD15066 | |
| PjCrus9 | 49 | 4.5 | 12.48 | 51.0 | BBC42585 | |
| Pandalus japonicus | PandjCrus IIb | 75 | 6.4 | 4.03 | 36.0 | AGU01544 |
| PandjCrus IIc | 21 | 2.1 | 12 | 23.8 | AGU01545 | |
| Panulirus japonicus | PanujCrus1 | 30 | 2.8 | 6.26 | 36.7 | ACU25382 |
| PanujCrus2 | 38 | 3.6 | 6.92 | 36.8 | ACU25383 | |
| PanujCrus3 | 32 | 3.3 | 5.74 | 31.2 | ACU25384 | |
| Penaeus setiferus | PseCrus1 | 22 | 1.99 | 9.75 | 27.3 | AAL36896 |
| PseCrus2 | 86 | 6.7 | 8.6 | 62.8 | AAL36897 | |
| Penaeus subtilis | PsuCrus1 | 44 | 3.7 | 8.75 | 54.5 | ABO93323 |
| Penaeus brasiliensis | PbCrus1 | 72 | 6.0 | 8.75 | 55.6 | ABQ96197 |
| Penaeus paulensis | PpCrus1 | 66 | 5.4 | 11 | 56.1 | ABM63361 |
| Penaeus chinensis | PcCrus2 | 28 | 2.76 | 12 | 25.0 | ACZ43782 |
| Penaeus schmitti | PscCrus1 | 64 | 5.2 | 8.6 | 57.8 | ABM63362 |
| Penaeus indicus | PiCrus1 | 33 | 3.1 | 4.65 | 33.3 | ACV84092 |
| Paralithodes camtschaticus | PcamCrus1 | 26 | 2.6 | 3.56 | 19.2 | ACJ06765 |
| Primers | Sequence (5′–3′) |
|---|---|
| Real-time PCR | |
| SpCrus2RF | TAGACGGACTTACTGGGAGCAT |
| SpCrus2RR | GGGACATTCACACCGCACT |
| 18s rRNA | |
| 18SRF | CAGACAAATCGCTCCACCAAC |
| 18SRR | GACTCAACACGGGGAACCTCA |
| RNAi | |
| SpCrus2iF | GCGTAATACGACTCACTATAGGGGATGACACCAGGGGGGGCT |
| SpCrus2iR | GCGTAATACGACTCACTATAGGGTTAGGCGGAAGGTTTACAC |
| EGFPiF | GCGTAATACGACTCACTATAGGGTGGTCCCAATTCTCGTGGAC |
| EGFPiR | GCGTAATACGACTCACTATAGGGCTTGAAGTTGACCTTGATGCC |
| Protein expression | |
| VP15EF | TACTCAGAATTCATGACAAAATACCCCGAGAT |
| VP15ER | TACTCACTCGAGTTAACGCCTTGACTTGCGGAC |
| VP19EF | TACTCAGAATTCATGGCCACCACGACTAACAC |
| VP19ER | TACTCACTCGAGATCCCTGGTCCTGTTCTTAT |
| VP26EF | TACTCAGGATCCACACGTGTTGGAAGAAGCGT |
| VP26ER | TACTCACTCGAGCTTCTTCTTGATTTCGTCCT |
| Co-immunoprecipitation (Co-IP) | |
| SpCrus2CF | CGGAATTCGCCATGGACACCAGGGGGGGCTTCCAC |
| SpCrus2CR | CCGCTCGAGCGGGCGGAAGGTTTACACACGGGATGG |
| VP26CF | GGGGTACCGCCACCATGGGTGTTGGAAGAAGCGTC |
| VP26CR | GCTCTAGACTCTTCTTCTTGATTTCGTCCTTG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhang, C.; Fang, W.-H.; Ma, H.-Y.; Li, X.-C. SpCrus2 Glycine-Rich Region Contributes Largely to the Antiviral Activity of the Whole-Protein Molecule by Interacting with VP26, a WSSV Structural Protein. Mar. Drugs 2021, 19, 544. https://doi.org/10.3390/md19100544
Wang Y, Zhang C, Fang W-H, Ma H-Y, Li X-C. SpCrus2 Glycine-Rich Region Contributes Largely to the Antiviral Activity of the Whole-Protein Molecule by Interacting with VP26, a WSSV Structural Protein. Marine Drugs. 2021; 19(10):544. https://doi.org/10.3390/md19100544
Chicago/Turabian StyleWang, Yue, Chao Zhang, Wen-Hong Fang, Hong-Yu Ma, and Xin-Cang Li. 2021. "SpCrus2 Glycine-Rich Region Contributes Largely to the Antiviral Activity of the Whole-Protein Molecule by Interacting with VP26, a WSSV Structural Protein" Marine Drugs 19, no. 10: 544. https://doi.org/10.3390/md19100544





