Chemical and Pharmacological Prospection of the Ascidian Cystodytes dellechiajei
Abstract
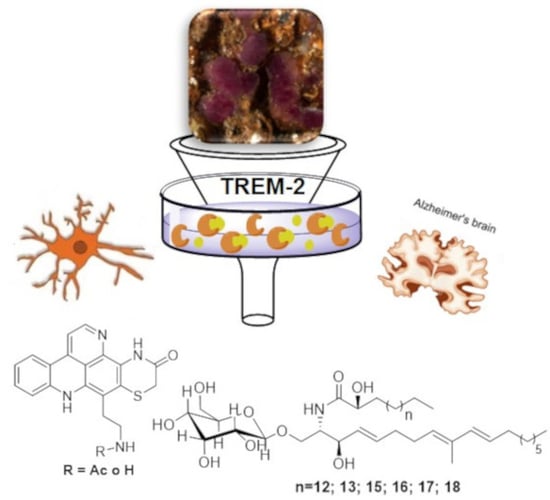
:1. Introduction
2. Results
2.1. Identification of the Colonies
2.2. Screening Platform and Bioactive Molecule Isolation
2.3. Isolation and Structure Characterization of the Bioactive Molecules
2.4. Biological Activity
3. Discussion
4. Materials and Methods
4.1. General Experimental Procedures
4.2. Sample Collection and Molecular Identification
4.3. Extraction and Isolation of Metabolites
4.4. TREM2 Reporter Assay
4.5. Cytotoxicity Assay
4.6. Binding Activity of TLR2, TLR4, and Dectin-1b
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Li, Y.; Sun, Y.; Wang, W.; Song, X.; Zhang, D. Chemical diversity and biological activities of marine-derived sulphur containing alkaloids: A comprehensive update. Arab. J. Chem. 2023, 16, 105011. [Google Scholar] [CrossRef]
- Guo, F.-W.; Zhang, Q.; Gu, Y.-C.; Shao, C.-L. Sulfur-containing marine natural products as leads for drug discovery and development. Curr. Opin. Chem. Biol. 2023, 75, 102330. [Google Scholar] [CrossRef]
- Rangel, K.C.; Debonsi, H.M.; Clementino, L.C.; Graminha, M.A.S.; Vilela, L.Z.; Colepicolo, P.; Gaspar, L. Antileishmanial activity of the red algae Iridaea cordata (Gigartinaceae; Rhodophyta). J. Appl. Phycol. 2019, 31, 825–834. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, G.A. Pyridoacridine alkaloids from deep-water marine organisms: Structural elucidation. Bull. Fac. Pharm. 2016, 54, 107–135. [Google Scholar] [CrossRef]
- Martínez-Garcia, M.; Díaz-Valdés, M.; Ramos-Esplá, A.; Salvador, N.; Lopez, P.; Larriba, E.; Antón, J. Cytotoxicity of the Ascidian Cystodytes dellechiajei against tumor cells and study of the involvement of associated microbiota in the production of cytotoxic compounds. Mar. Drugs 2007, 5, 52–70. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.-H.; Wang, Y.-W.; Yang, J.; Tong, Z.-J.; Wu, Z.-J.; Wang, Y.-B.; Wang, Q.-X.; Li, Q.-Q.; Yu, Y.-C.; Leng, X.-J.; et al. Natural products as potential lead compounds to develop new antiviral drugs over the past decade. Eur. J. Med. Chem. 2023, 260, 115726. [Google Scholar] [CrossRef]
- López-Legentil, S.; Turon, X. How do morphotypes and chemotypes relate to genotypes? The colonial ascidian Cystodytes (Polycitoridae). Zool. Scr. 2005, 34, 3–14. [Google Scholar] [CrossRef]
- López-Legentil, S.; Turon, X. Population genetics, phylogeography and speciation of Cystodytes (Ascidiacea) in the western Mediterranean Sea. Biol. J. Linn. Soc. 2006, 88, 203–214. [Google Scholar] [CrossRef]
- López-Legentil, S.; Ruchty, M.; Domenech, A.; Turon, X. Life cycles and growth rates of two morphotypesof Cystodytes (Ascidiacea) in the west Mediterranean. Mar. Ecol. Prog. Ser. 2005, 296, 219–228. [Google Scholar] [CrossRef]
- Moreiras-Figueruelo, A.; Nuzzo, G.; Galasso, C.; Sansone, C.; Crocetta, F.; Mazella, V.; Gallo, C.; Barra, G.; Sardo, A.; Luliano, A.; et al. Probing the therapeutic potential of marine phyla by SPE extraction. Mar. Drugs 2021, 19, 640. [Google Scholar] [CrossRef]
- Gallo, C.; Barra, G.; Saponaro, M.; Manzo, E.; Fioretto, L.; Ziaco, M.; Nuzzo, G.; D’Ippolito, G.; De Palma, R.; Fontana, A. A new bioassay platform design for the discovery of small molecules with anticancer immunotherapeutic activity. Mar. Drugs 2020, 18, 604. [Google Scholar] [CrossRef]
- Mecca, C.; Giambanco, I.; Donato, R.; Arcuri, C. Microglia and aging: The role of the TREM2-DAP12 and CX3CL1-CX3CR1 axes. Int. J. Mol. Sci. 2018, 19, 318. [Google Scholar] [CrossRef]
- Perugorria, M.J.; Esparza-Baquer, A.; Oakley, F.; Labiano, I.; Korosec, A.; Jais, A.; Mann, J.; Tiniakos, D.; Santos-Laso, A.; Arbelaiz, A.; et al. Non-parenchymal TREM-2 protects the liver from immune-mediated hepatocellular damage. Hepatology 2019, 68, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Piccio, L.; Buonsanti, C.; Cella, M.; Tassi, I.; Schmidt, R.E.; Fenoglio, C.; Rinker, J., II; Naismith, R.T.; Panina-Bordignon, P.; Passini, N.; et al. Identification of soluble TREM-2 in the cerebrospinal fluid and its association with multiple sclerosis and CNS inflammation. Brain 2008, 131, 3081–3091. [Google Scholar] [CrossRef] [PubMed]
- Ulland, T.K.; Song, W.M.; Huang, S.C.; Ulrich, J.D.; Sergushichev, A.; Beatty, W.L.; Loboda, A.A.; Zhou, Y.; Cairns, N.J.; Kambal, A.; et al. TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell 2017, 170, 649–663. [Google Scholar] [CrossRef] [PubMed]
- Hamerman, J.A.; Jarjoura, J.R.; Humphrey, M.B.; Nakamura, M.C.; Seaman, W.E.; Lanier, L.L. Cutting edge: Inhibition of TRL and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J. Immunol. 2006, 177, 2051–2055. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, I.R.; Gilfillan, S.; Cella, M.; Aoshi, T.; Miller, M.; Piccio, L.; Hernandez, M.; Collona, M. Cutting edge: TREM-2 attenuates macrophage activation. J. Immunol. 2006, 177, 3520–3524. [Google Scholar] [CrossRef]
- Zhong, L.; Zhang, Z.-L.; Li, X.; Liao, C.; Mou, P.; Wang, T.; Wang, Z.; Wei, M.; Xu, H.; Bu, G.; et al. TREM2/DAP12 complex regulates inflammatory responses in microglia via the JNK signaling pathway. Front. Aging Neurosci. 2017, 9, 204. [Google Scholar] [CrossRef]
- Gallo, C.; Manzo, E.; Barra, G.; Fioretto, L.; Ziaco, M.; Nuzzo, G.; D’Ippolito, G.; Ferrera, F.; Contini, P.; Castiglia, D.; et al. Sulfavant A as the first synthetic TREM2 ligand discloses a homeostatic response of dendritic cells after receptor angagement. Cell. Mol. Life Sci. 2022, 79, 369. [Google Scholar] [CrossRef]
- Cutignano, A.; Nuzzo, G.; Ianora, A.; Luongo, E.; Romano, G.; Gallo, C.; Sansone, C.; Aprea, S.; Mancini, F.; D’Oro, U.; et al. Development and application of a novel SPE-method for bioassay-guided fractionation of marine extracts. Mar. Drugs 2015, 13, 5736–5749. [Google Scholar] [CrossRef]
- Nuzzo, G.; Gallo, C.; Crocetta, F.; Romano, L.; Barra, G.; Senese, G.; Dell’Isola, M.; Carbone, D.; Tanduo, V.; Albiani, F.; et al. Identification of marine alkaloid Lepadin as a potential inducer of immunogenic cell death. Biomolecules 2022, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Lizasa, E.; Chuma, Y.; Uematsu, T.; Kubota, M.; Kawaguchi, H.; Umemura, M.; Toyonaga, K.; Kiyohara, H.; Yano, I.; Colonna, M.; et al. TREM2 is a receptor for non-glycosilated mycolic acids of mycobacteria that limits anti-mycobacterial macrophage activation. Nat. Commun. 2021, 12, 2299. [Google Scholar]
- Carroll, A.R.; Cooray, N.M.; Poiner, A.; Scheuer, P.J. A secondary shermilamine alkaloid from a tunicate Trididemnum sp. J. Org. Chem. 1989, 54, 4232–4235. [Google Scholar] [CrossRef]
- Bontemps, N.; Bry, D.; López-Legentil, S.; Simon-Levert, A.; Long, C.; Banaigs, B. Structures and antimicrobial activities of pyridoacridines alkaloids isolated from different chromotypes of the Ascidian Cystodytes dellechiajei. J. Nat. Prod. 2010, 73, 1044–1048. [Google Scholar] [CrossRef]
- Careaga, V.P.; Maier, M.S. Cerebrosides from marine organisms. Stud. Nat. Prod. Chem. 2014, 42, 59–81. [Google Scholar]
- Cheng, S.-Y.; Wen, Z.-H.; Chiou, S.-F.; Tsai, C.-W.; Wang, S.-K.; Hsu, C.-H.; Dai, C.-F.; Chiang, M.Y.; Wang, W.-H.; Duh, C.-Y. Ceramide and cerebrosides from the octocoral Sarcophyton ehrenbergi. J. Nat. Prod. 2009, 72, 465–468. [Google Scholar] [CrossRef]
- Maier, M.S.; Kuriss, A.; Seldes, A.M. Isolation and structure of glucosylceramides from the starfish Cosmasterias lurida. Lipids 1998, 33, 825–827. [Google Scholar] [CrossRef]
- de Vivar, M.E.D.; Seldes, A.M.; Maier, S.M. Two novel glucosylceramides from gonads and body walls of the Patagonian starfish Allostichaster inaequalis. Lipids 2002, 37, 597–603. [Google Scholar] [CrossRef]
- Jin, W.; Rinehart, K.L.; Jares-Erijman, E.A. Ophidiacerebrosides: Cytotoxic glicosphingolipids containing a novel sphingosine from sea star. J. Org. Chem. 1994, 59, 144–147. [Google Scholar] [CrossRef]
- Filipello, F.; Goldsbury, C.; You, S.F.; Locca, A.; Karch, C.M.; Piccio, L. Soluble TREM-2: Innocent or active player in neurological diseases? Neurobiol. Dis. 2022, 165, 105630. [Google Scholar] [CrossRef]
- Labiano, I.; Agirre-Lizaso, A.; Olaizola, P.; Echebarria, A.; Huici-Izagirre, M.; Olaizola, I.; Esparza-Baquer, A.; Shariff, O.; Hijona, E.; Milkiewicz, P.; et al. TREM-2 plays a protective role in cholestasis by acting as a negative regulator of inflammation. J. Hepatol. 2022, 77, 991–1004. [Google Scholar] [CrossRef]
- Aiello, A.; Fattorusso, E.; Mangoni, A.; Menna, M. Three new 2,3-dihydroxi fatty acid glycosphingolipids from the Mediterranean tunicate Microcosmus sulcatus. Eur. J. Org. Chem. 2003, 4, 734–739. [Google Scholar] [CrossRef]
- Loukaci, A.; Bultel-Poncé, V.; Longeon, A.; Guyot, M. New lipids from the tunicate Cystodytes dellechiajei, as PLA2 inhibitors. J. Nat. Prod. 2000, 63, 799–802. [Google Scholar] [CrossRef]
- Carroll, A.R.; Scheuer, P.J. Kuanoniamines A, B, C and D: Pentacyclic alkaloids from a tunicate and its prosobranch mollusk predator Chelynotus semperi. J. Org. Chem. 1990, 55, 4427–4431. [Google Scholar] [CrossRef]
- Delfourne, E.; Bastide, J. Marine Pyridoacridine alkaloids and synthetic analogues as antitumor agents. Med. Res. Rev. 2003, 23, 234–252. [Google Scholar] [CrossRef] [PubMed]
- Bontemps, N.; Gattacceca, F.; Long, C.; Thomas, O.P.; Banaigs, B. Additional cytotoxic Pyridoacridine alkaloids from the Ascidian Cystodytes violatinctus and biogenetic considerations. J. Nat. Prod. 2013, 76, 1801–1805. [Google Scholar] [CrossRef] [PubMed]
- Bry, D.; Banaigs, B.; Long, C.; Bontemps, N. New pyridoacridine alkaloids from the purple morph of the Ascidian Cystodytes dellechiajei. Tetrahedron Lett. 2011, 52, 3041–3044. [Google Scholar] [CrossRef]
- Lindsay, B.S.; Pearce, A.N.; Copp, B.R. Efficient and convenient pyridine ring-e formation of the cytotoxic marine alkaloid Ascididemin and related analogues. Synth. Commun. 1997, 27, 2587–2592. [Google Scholar] [CrossRef]
- Eder, C.; Schupp, P.; Proksch, P.; Wray, V.; Steube, K.; Muller, C.E.; Frobenius, W.; Herderich, M.; van Soest, R.W.M. Bioactive Pyridoacridine alkaloids from the Micronesian sponge Oceanapia sp. J. Nat. Prod. 1998, 61, 301–305. [Google Scholar] [CrossRef]
- Kobayashi, J.; Cheng, J.-F.; Walchli, M.R.; Nakamura, H.; Hirata, Y.; Sasaki, T.; Ohizumi, Y. Cystodins A, B and C, novel tetracyclic aromatic alkaloids with potent antineoplastic activity from the Okinawan tunicate Cystodytes dellechiajei. J. Org. Chem. 1988, 53, 1800–1804. [Google Scholar] [CrossRef]
- Ibrahim, S.R.M.; Mohamed, G.A.; Elkhayat, E.S.; Fouad, M.A.; Proksch, P. Sagitol C, a new cytotoxic pyridoacridine alkaloid from the sponge Oceanapia sp. Bull. Fac. Pharm. 2013, 51, 229–232. [Google Scholar] [CrossRef]
- Feng, Y.; Davis, R.A.; Sykes, M.L.; Avery, V.M.; Carroll, A.R.; Camp, D.; Quinn, R.J. Antitrypanosomal pyridoacridine alkaloids from the Australian ascidian Polysyncraton echinatum. Tetrahedron Lett. 2010, 51, 2477–2479. [Google Scholar] [CrossRef]
- Virgili, R.; Tanduo, V.; Katsanevakis, S.; Terlizzi, F.; Villani, G.; Fontana, A.; Crocetta, F. The Miseno lake (central-western Mediterranean Sea): An overlooked reservoir of non-indigenous and cryptogenic Ascidians in marine reserve. Front. Mar. Sci. 2022, 19, 866906. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; Van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-Tree: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Rambaut, A.; Drummond, A.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef]
- Del Prete, E.; Campos, A.M.; Rocca, F.D.; Gallo, C.; Fontana, A.; Nuzzo, G.; Angelini, C. ADViSELipidomics: A workflow for analyzing lipidomics data. Bioinformatics 2022, 38, 5460–5462. [Google Scholar] [CrossRef]






| 1 | 2 | ||||||
|---|---|---|---|---|---|---|---|
| No. | δH a (mult, J in Hz) | δH a (mult, J in Hz) | δCa | δH b (mult, J in Hz) | δCb | δHd (mult, J in Hz) | δCd |
| 2 | 8.55 (d, 5.1) | 8.54 (d, 5.0) | 151.5 | 8.55 (d, 5.5) | 150.8 | 8.55 (d, 5.3) | 150.5 |
| 3 | 7.50 (d, 5.1) | 7.49 (d, 5.0) | 108.8 | 7.56 (d,5.2) | 107.7 | 7.63 (d, 5.3) | 107.3 |
| 3a | 141.8 | 140.3 | 142.5 | ||||
| 3b | 117.4 | 115.5 | 115.4 | ||||
| 4 | 8.02 (d, 8.2) | 8.00 (d, 8.1) | 125.1 | 8.05 (d, 8.1) | 124.1 | 8.14 (d, 8.1) | 125.0 |
| 5 | 7.11 (m) | 7.09 (t, 7.8) | 123.0 | 7.04 (d, 7.9) | 121.3 | 7.13 (d, 7.9) | 122.2 |
| 6 | 7.46 (overlapped) | 7.44 (t, 7.8) | 133.3 | 7.43 (d, 7.9) | 131.5 | 7.53 (t, 7.9) | 132.7 |
| 7 | 7.45 (overlapped) | 7.53 (d, 8.1) | 118.0 | 7.73 (d, 8.1) | 116.7 | 7.48 (d, 8.1) | 116.9 |
| 7a | 141.4 | 139.8 | 140.0 | ||||
| 8a | 133.6 | 131.9 | 131.5 | ||||
| 9 | 107.3 | 107.8 | 108.4 | ||||
| 9a | 123.5 | 124.0 | 124.4 | ||||
| 11 | 3.55 (s) | 3.63 (s) | 30.5 | 3.61 (s) | 29.1 | 3.64 (s) | 29.4 |
| 12 | 166.5 | 163.8 | 164.0 | ||||
| 13a | 123.3 | 121.9 | 121.7 | ||||
| 13b | 139.4 | 137.2 | 136.0 | ||||
| 13c | 118.6 | 116.9 | 117.1 | ||||
| 14 | 3.33 (overlapped) | 3.33 (t, 8) | 28.1 | 3.24 (t, 7.5) | 27.1 c | 3.21 (t, 8.1) | 25.8 |
| 15 | 3.14 (bt) | 3.14 (t, 8) | 38.7 | 2.91 (t, 7.5) | 37.3 c | 2.98 (brt) | 36.9 |
| 18 | 1.92 (s) | - | - | ||||
| 3 | ||
|---|---|---|
| No. | δH (mult, J in Hz) | δC |
| 1 | 3.73 (dd, 3.6; 10.3) 4.14 (dd, 4.7; 10.6) | 69.7 |
| 2 | 4.02 (m) | 54.6 |
| 3 | 4.17 (bt, 7.4) | 73.1 |
| 4 | 5.53 (dd, 7.5; 15.2) | 131.3 |
| 5 | 5.77 (dt, 6.4; 15.4) | 134.3 |
| 6 | 2.12 (m) | 34.0 |
| 7 | 2.24 (m) | 30.4 |
| 8 | 5.38 (t, 7.2) | 130.3 |
| 10 | 6.01 (d, 15.8) | 136.0 |
| 11 | 5.58 (dt, 6.8; 15.5) | 128.6 |
| 12 | 2.11 (m) | 33.5 |
| 13–17 | 1.37–1.30 | 23.8–31.0 |
| 18 | 0.94–0.91 | 14.5 |
| 19 | 1.74 (s) | 12.0 |
| 2′ | 4.02 (m) | 73.1 |
| 3′ | 1.57 (m) | 36.0 |
| (CH2)n | 1.37–1.30 | 23.8–31.0 |
| CH3 | 0.94–0.91 | 14.5 |
| 1″ | 4.29 (d, 7.82) | 104.8 |
| 2″ | 3.23 (m) | 75.0 |
| 3″ | 3.38 (m) | 77.9 |
| 4″ | 3.30 (m) | 71.6 |
| 5″ | 3.31 (m) | 78.0 |
| 6″ | 3.90 (d, 11.9) 3.70 (dd, 4.12; 11.2) | 62.7 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batista, P.J.; Nuzzo, G.; Gallo, C.; Carbone, D.; dell’Isola, M.; Affuso, M.; Barra, G.; Albiani, F.; Crocetta, F.; Virgili, R.; et al. Chemical and Pharmacological Prospection of the Ascidian Cystodytes dellechiajei. Mar. Drugs 2024, 22, 75. https://doi.org/10.3390/md22020075
Batista PJ, Nuzzo G, Gallo C, Carbone D, dell’Isola M, Affuso M, Barra G, Albiani F, Crocetta F, Virgili R, et al. Chemical and Pharmacological Prospection of the Ascidian Cystodytes dellechiajei. Marine Drugs. 2024; 22(2):75. https://doi.org/10.3390/md22020075
Chicago/Turabian StyleBatista, Pedro Jatai, Genoveffa Nuzzo, Carmela Gallo, Dalila Carbone, Mario dell’Isola, Mario Affuso, Giusi Barra, Federica Albiani, Fabio Crocetta, Riccardo Virgili, and et al. 2024. "Chemical and Pharmacological Prospection of the Ascidian Cystodytes dellechiajei" Marine Drugs 22, no. 2: 75. https://doi.org/10.3390/md22020075