Study on the Anti-Mycobacterium marinum Activity of a Series of Marine-Derived 14-Membered Resorcylic Acid Lactone Derivatives
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
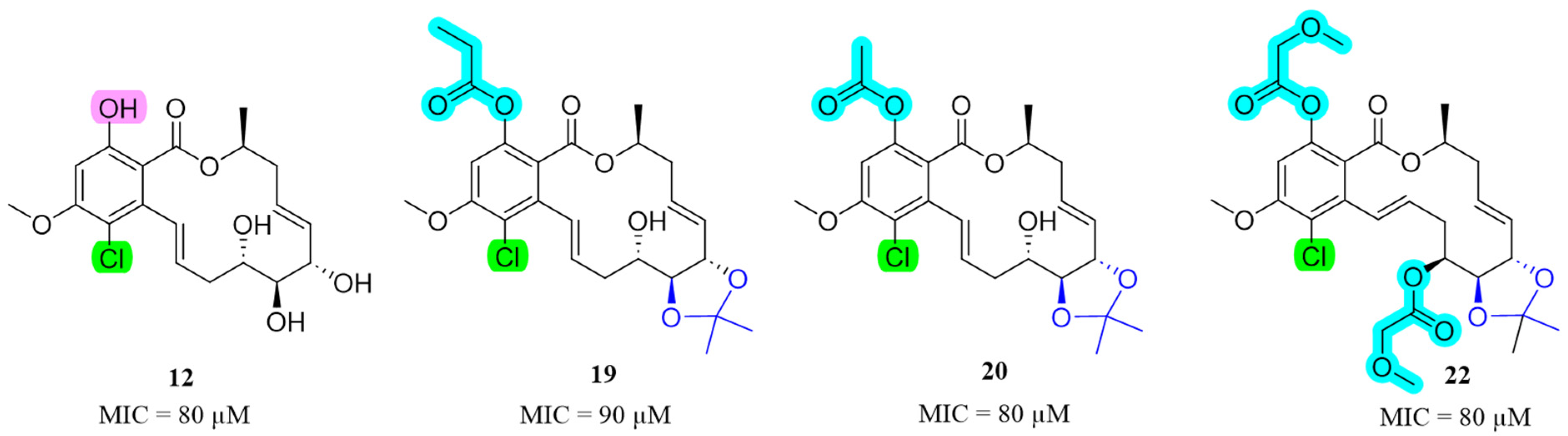
2.2. Evaluation of Biological Activity
2.2.1. Anti-M. marinum and Other Antimicrobial Activity
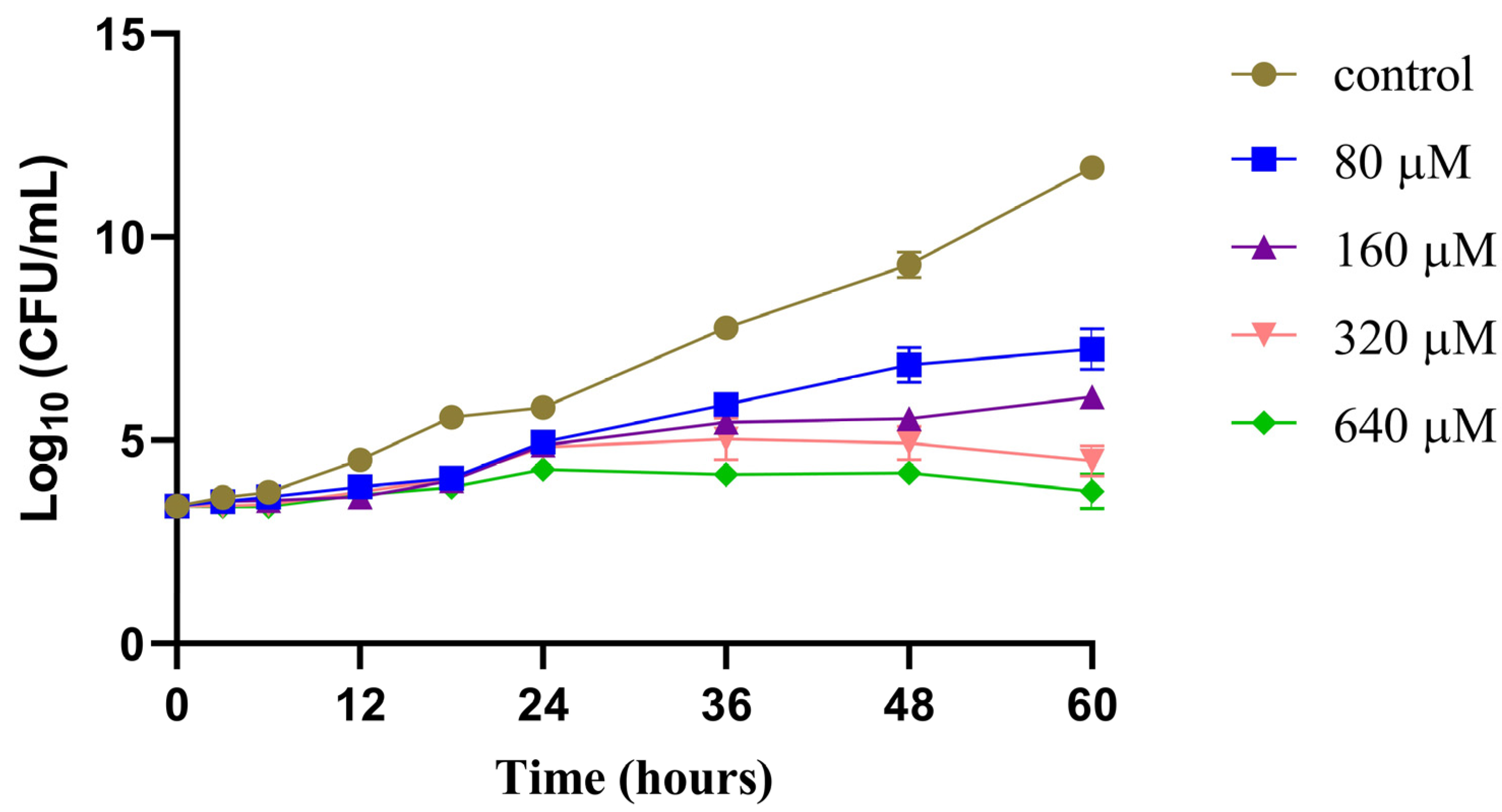
2.2.2. Time−Growth Curves of M. marinum Strain Treated with Different Concentrations of Compound 12
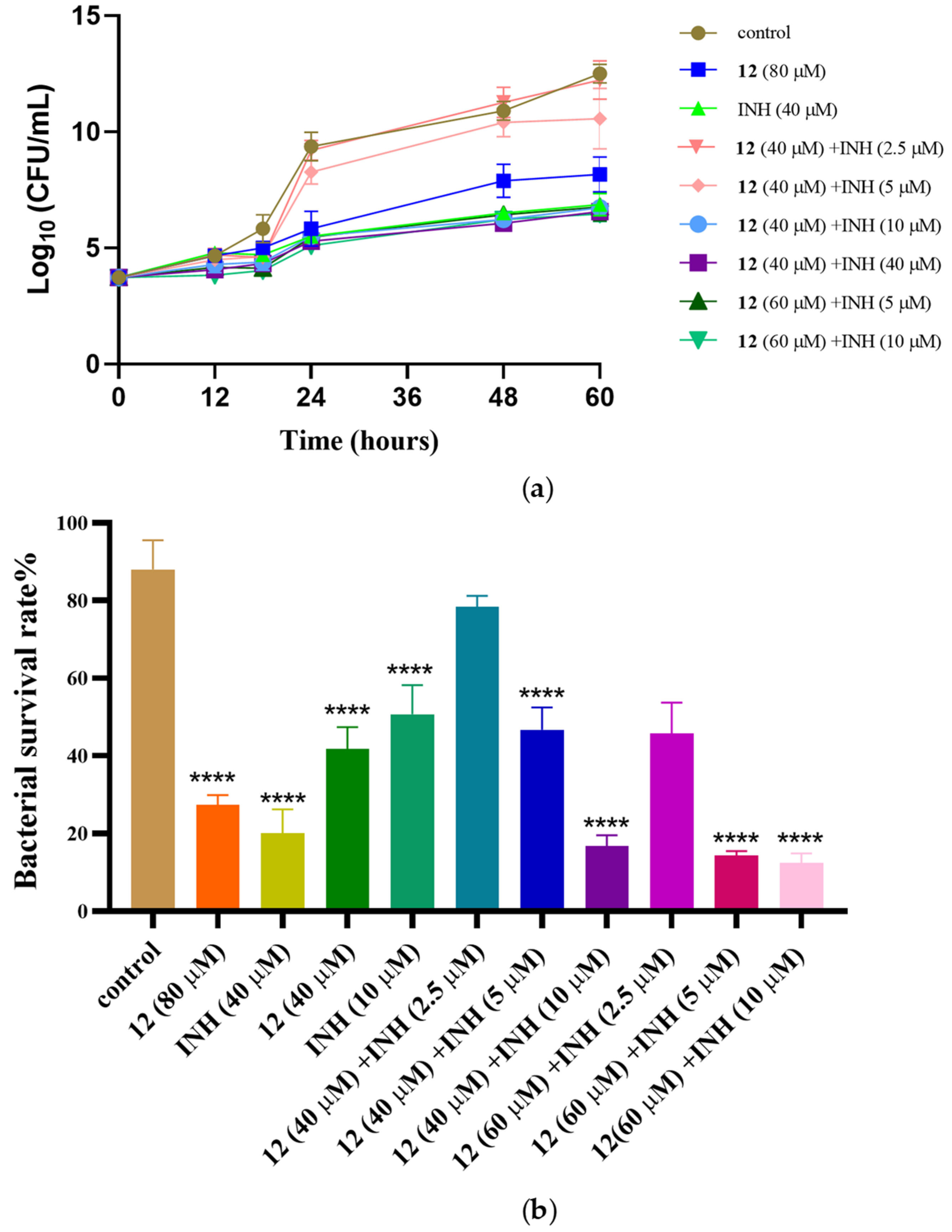
2.2.3. Anti-M. marinum Effects of Compound 12 in Combination with Positive Drugs
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fungal Material
3.3. Fermentation, Extraction and Isolation
3.4. General Synthetic Methods for Compounds 2–97
3.4.1. General Procedure for the Synthesis of 19
3.4.2. General Procedure for the Synthesis of 24–38
3.4.3. Characterization Data of Compounds 19, and 24–38
3.5. Antimicrobial Activity
3.6. Time–Growth Curve Assay
3.7. In Vitro Synergic Anti-M. marinum Activity Assay
3.8. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Daniel, T.M. The History of Tuberculosis. Respir. Med. 2006, 100, 1862–1870. [Google Scholar] [CrossRef]
- Public Health Online. Available online: https://www.publichealthonline.org/worst-global-pandemics-in-history (accessed on 13 October 2021).
- Global Tuberculosis Report 2023. World Health Organization: Geneva, Switzerland, 2023. Licence: CC BY-NC-SA 3.0 IGOnization. Available online: https://www.who.int/publications/i/item/9789240083851 (accessed on 7 November 2023).
- Bloom, B.R. A half-century of research on tuberculosis: Successes and challenges. J. Exp. Med. 2023, 220, e20230859. [Google Scholar] [CrossRef] [PubMed]
- Maitre, T.; Aubry, A.; Jarlier, V.; Robert, J.; Veziris, N.; Bernard, C.; Sougakoff, W.; Brossier, F.; Cambau, E.; Mougari, F.; et al. Multidrug and Extensively Drug-Resistant Tuberculosis. Med. Mal. Infect. 2017, 47, 3–10. [Google Scholar] [CrossRef] [PubMed]
- Lange, C.; Dheda, K.; Chesov, D.; Mandalakas, A.M.; Udwadia, Z.; Horsburgh, C.R., Jr. Management of drug-resistant tuberculosis. Lancet 2019, 394, 953–966. [Google Scholar] [CrossRef] [PubMed]
- Elsevier. Patient safety: Too little, but not too late. Lancet 2019, 394, 895. [Google Scholar] [CrossRef]
- Kakkar, A.K.; Dahiya, N. Bedaquiline for the treatment of resistant tuberculosis: Promises and pitfalls. Tuberculosis 2014, 94, 357–362. [Google Scholar] [CrossRef] [PubMed]
- Ryan, N.J.; Lo, J.H. Delamanid: First global approval. Drugs 2014, 74, 1041–1045. [Google Scholar] [CrossRef]
- Keam, S.J. Pretomanid: First Approval. Drugs 2019, 79, 1797–1803. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, G.Z.; Xu, M. Biohazard levels and biosafety protection for Mycobacterium tuberculosis strains with different virulence. Biosaf. Health 2020, 2, 135–141. [Google Scholar] [CrossRef]
- van Soolingen, D.; Wisselink, H.J.; Lumb, R.; Anthony, R.; van der Zanden, A.; Gilpin, C. Practical biosafety in the tuberculosis laboratory: Containment at the source is what truly counts. Int. J. Tuberc. Lung Dis. 2014, 18, 885–889. [Google Scholar] [CrossRef]
- Lambrecht, R.S.; Carriere, J.F.; Collins, M.T. A model for analyzing growth kinetics of a slowly growing Mycobacterium sp. Appl. Environ. Microbiol. 1988, 54, 910–916. [Google Scholar] [CrossRef]
- Gao, L.Y.; Groger, R.; Cox, J.S.; Beverley, S.M.; Lawson, E.H.; Brown, E.J. Transposon mutagenesis of Mycobacterium marinum identifies a locus linking pigmentation and intracellular survival. Infect. Immun. 2003, 71, 922–929. [Google Scholar] [CrossRef]
- Cronin, R.M.; Ferrell, M.J.; Cahir, C.W.; Champion, M.M.; Champion, P.A. Proteo-genetic analysis reveals clear hierarchy of ESX-1 secretion in Mycobacterium marinum. Proc. Natl. Acad. Sci. USA 2022, 119, e2123100119. [Google Scholar] [CrossRef] [PubMed]
- Van Seymortier, P.; Verellen, K.; De Jonge, I. Mycobacterium marinum causing tenosynovitis. ‘Fish tank finger’. Acta. Orthop. Belg. 2004, 70, 279–282. [Google Scholar] [PubMed]
- Stinear, T.P.; Seemann, T.; Harrison, P.F.; Jenkin, G.A.; Davies, J.K.; Johnson, P.D.; Abdellah, Z.; Arrowsmith, C.; Chillingworth, T.; Churcher, C.; et al. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res. 2008, 18, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Habjan, E.; Ho, V.Q.T.; Gallant, J.; van Stempvoort, G.; Jim, K.K.; Kuijl, C.; Geerke, D.P.; Bitter, W.; Speer, A. An anti-tuberculosis compound screen using a zebrafish infection model identifies an aspartyl-tRNA synthetase inhibitor. Dis. Model. Mech. 2021, 14, dmm049145. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Hai, Y.; Cai, Z.M.; Li, P.J.; Wei, M.Y.; Wang, C.Y.; Gu, Y.C.; Shao, C.L. Trends of antimalarial marine natural products: Progresses, challenges and opportunities. Nat. Prod. Rep. 2022, 39, 969–990. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2022, 39, 1122–1171. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Liu, X.; Zhang, L.; Quinn, R.J.; Feng, Y. Anti-mycobacterial natural products and mechanisms of action. Nat. Prod. Rep. 2022, 39, 77–89. [Google Scholar] [CrossRef]
- Shao, C.L.; Wu, H.X.; Wang, C.Y.; Liu, Q.A.; Xu, Y.; Wei, M.Y.; Qian, P.Y.; Gu, Y.C.; Zheng, C.J.; She, Z.G.; et al. Potent antifouling resorcylic acid lactones from the gorgonian-derived fungus Cochliobolus lunatus. J. Nat. Prod. 2011, 74, 629–633. [Google Scholar] [CrossRef] [PubMed]
- Jana, N.; Nanda, S. Resorcylic acid lactones (RALs) and their structural congeners: Recent advances in their biosynthesis, chemical synthesis and biology. New J. Chem. 2018, 42, 17803–17873. [Google Scholar] [CrossRef]
- Liu, Q.A.; Shao, C.L.; Gu, Y.C.; Blum, M.; Gan, L.S.; Wang, K.L.; Chen, M.; Wang, C.Y. Antifouling and Fungicidal Resorcylic Acid Lactones from the Sea Anemone-Derived Fungus Cochliobolus lunatus. J. Agric. Food. Chem. 2014, 62, 3183–3191. [Google Scholar] [CrossRef]
- Xu, W.F.; Xue, X.J.; Qi, Y.X.; Wu, N.N.; Wang, C.Y.; Shao, C.L. Cochliomycin G, a 14-membered resorcylic acid lactone from a marine-derived fungus Cochliobolus lunatus. Nat. Prod. Res. 2021, 35, 490–493. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Shao, C.L.; Chen, M.; Liu, Q.A.; Wang, C.Y. Brominated resorcylic acid lactones from the marine-derived fungus Cochliobolus lunatus induced by histone deacetylase inhibitors. Tetrahedron Lett. 2014, 55, 4888–4891. [Google Scholar] [CrossRef]
- Wang, K.L.; Zhang, G.; Sun, J.; Xu, Y.; Han, Z.; Liu, L.L.; Shao, C.L.; Liu, Q.A.; Wang, C.Y.; Qian, P.Y. Cochliomycin A inhibits the larval settlement of Amphibalanus amphitrite by activating the NO/cGMP pathway. Biofouling 2016, 32, 35–44. [Google Scholar] [CrossRef]
- Zhang, X.Q.; Spadafora, C.; Pineda, L.M.; Ng, M.G.; Sun, J.H.; Wang, W.; Wang, C.Y.; Gu, Y.C.; Shao, C.L. Discovery, Semisynthesis, Antiparasitic and Cytotoxic Evaluation of 14-Membered Resorcylic Acid Lactones and Their Derivatives. Sci. Rep. 2017, 7, 11822. [Google Scholar] [CrossRef]
- Xu, W.F.; Wu, N.N.; Wu, Y.W.; Qi, Y.X.; Wei, M.Y.; Pineda, L.M.; Ng, M.G.; Spadafora, C.; Zheng, J.Y.; Lu, L.; et al. Structure modification, antialgal, antiplasmodial, and toxic evaluations of a series of new marine-derived 14-membered resorcylic acid lactone derivatives. Mar. Life Sci. Technol. 2022, 4, 88–97. [Google Scholar] [CrossRef]
- Koul, A.; Arnoult, E.; Lounis, N.; Guillemont, J.; Andries, K. The challenge of new drug discovery for tuberculosis. Nature 2011, 469, 483–490. [Google Scholar] [CrossRef]
- Smith, T.C., 2nd; Aldridge, B.B. Targeting drugs for tuberculosis. Science 2019, 364, 1234–1235. [Google Scholar] [CrossRef]
- Li, Z.; Huang, Y.; Tu, J.; Yang, W.; Liu, N.; Wang, W.; Sheng, C. Discovery of BRD4-HDAC Dual Inhibitors with Improved Fungal Selectivity and Potent Synergistic Antifungal Activity against Fluconazole-Resistant Candida albicans. J. Med. Chem. 2023, 66, 5950–5964. [Google Scholar] [CrossRef] [PubMed]
- Fromtling, R.A.; Galgiani, J.N.; Pfaller, M.A.; Espinel-Ingroff, A.; Bartizal, K.F.; Bartlett, M.S.; Body, B.A.; Frey, C.; Hall, G.; Roberts, G.D. Multicenter evaluation of a broth macrodilution antifungal susceptibility test for yeasts. Antimicrob. Agents Chemother. 1993, 37, 39–45. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Liu, N.; Tu, J.; Ji, C.; Han, G.; Sheng, C. Discovery of simplified sampangine derivatives with potent antifung alactivities against cryptococcal meningitis. ACS Infect. Dis. 2019, 5, 1376–1384. [Google Scholar] [CrossRef] [PubMed]

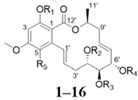 | |||||||||||
| No. | R1 | R2 | R3 | R4 | R5 | No. | R1 | R2 | R3 | R4 | R5 |
| 1 | H | H | H | H | H | 9 |  | H | H | H | Cl |
| 2 | 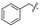 | H | H | H | H | 10 |  | H | H | H | Cl |
| 3 |  | H | H | H | H | 11 | 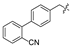 | H | H | H | Cl |
| 4 | 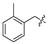 | H | H | H | H | 12 | H | H | H | H | Cl |
| 5 | 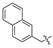 | H | H | H | H | 13 | 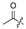 |  | 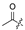 | 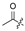 | Cl |
| 6 | 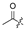 |  |  |  | H | 14 | 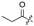 |  | 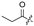 | 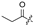 | Cl |
| 7 | 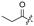 | 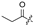 | 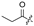 |  | H | 15 |  |  | H |  | Cl |
| 8 |  | 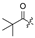 | H |  | H | 16 | 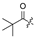 | H | H | H | Cl |
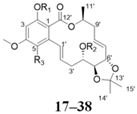 | |||||||||||
| No. | R1 | R2 | R3 | No. | R1 | R2 | R3 | ||||
| 17 | H | H | H | 28 | 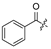 | H | H | ||||
| 18 | H | H | Cl | 29 | 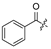 |  | H | ||||
| 19 | 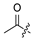 | H | Cl | 30 |  | H | H | ||||
| 20 | 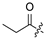 | H | Cl | 31 |  | 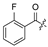 | H | ||||
| 21 |  | H | Cl | 32 | H |  | H | ||||
| 22 |  | 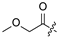 | Cl | 33 | 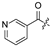 | H | H | ||||
| 23 | 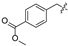 | H | Cl | 34 |  |  | H | ||||
| 24 | 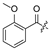 | H | H | 35 | H | 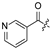 | H | ||||
| 25 | 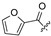 | H | H | 36 |  | H | H | ||||
| 26 | 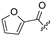 | 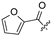 | H | 37 | 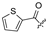 | H | H | ||||
| 27 | H |  | H | 38 |  |  | H | ||||
| Compound | MIC90 (µM) | |||||
|---|---|---|---|---|---|---|
| M. marinum | S. aureus | E. coli | P. aeruginosa | C. albicans | A. fumigatus | |
| 1 | >200 | >100 | >100 | >100 | >100 | >100 |
| 4 | >200 | 12.5 | >100 | >100 | >100 | >100 |
| 11 | 100 | >100 | >100 | >100 | >100 | >100 |
| 12 | 80 | >100 | >100 | >100 | >100 | >100 |
| 18 | >200 | >100 | >100 | >100 | >100 | >100 |
| 19 | 90 | >100 | >100 | >100 | >100 | >100 |
| 20 | 80 | >100 | >100 | >100 | >100 | >100 |
| 22 | 80 | >100 | >100 | >100 | >100 | >100 |
| Isoniazid | 40 | nt | nt | nt | nt | nt |
| Rifampicin | 10 | nt | nt | nt | nt | nt |
| Ciprofloxacin | nt | 3.13 | 0.10 | 1.56 | nt | nt |
| Amphotericin B | nt | nt | nt | nt | 0.84 | 0.07 |
| Positive Drugs MIC90 (µM) | Compound 12 MIC90 (µM) | FICI 1 | Mode of Action | |||
|---|---|---|---|---|---|---|
| Alone | Combined | Alone | Combined | |||
| Isoniazid | 40 | 10 | 80 | 40 | 0.75 | additive |
| Rifampicin | 10 | 2.5 | 80 | 40 | 0.75 | additive |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jing, Q.-Q.; Yin, J.-N.; Cheng, Y.-J.; Zhang, Q.; Cao, X.-Z.; Xu, W.-F.; Shao, C.-L.; Wei, M.-Y. Study on the Anti-Mycobacterium marinum Activity of a Series of Marine-Derived 14-Membered Resorcylic Acid Lactone Derivatives. Mar. Drugs 2024, 22, 135. https://doi.org/10.3390/md22030135
Jing Q-Q, Yin J-N, Cheng Y-J, Zhang Q, Cao X-Z, Xu W-F, Shao C-L, Wei M-Y. Study on the Anti-Mycobacterium marinum Activity of a Series of Marine-Derived 14-Membered Resorcylic Acid Lactone Derivatives. Marine Drugs. 2024; 22(3):135. https://doi.org/10.3390/md22030135
Chicago/Turabian StyleJing, Qian-Qian, Jun-Na Yin, Ya-Jie Cheng, Qun Zhang, Xi-Zhen Cao, Wei-Feng Xu, Chang-Lun Shao, and Mei-Yan Wei. 2024. "Study on the Anti-Mycobacterium marinum Activity of a Series of Marine-Derived 14-Membered Resorcylic Acid Lactone Derivatives" Marine Drugs 22, no. 3: 135. https://doi.org/10.3390/md22030135






