Cytotoxic and Antibacterial Meroterpenoids Isolated from the Marine-Derived Fungus Talaromyces sp. M27416
Abstract
:1. Introduction
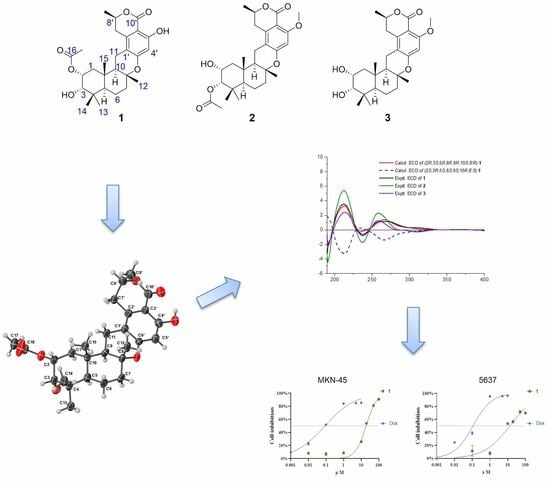
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Fermentation, Extraction, and Isolation
3.3. NMR Calculation
3.4. Cytotoxic and Antibacterial Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Newman, D.J.; Cragg, G.M. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J. Nat. Prod. 2020, 83, 770–803. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.R.; Copp, B.R.; Davis, R.A.; Keyzers, R.A.; Prinsep, M.R. Marine natural products. Nat. Prod. Rep. 2021, 38, 362–413. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.R.; Gong, L.Q.; Jin, M.Y.; Wang, R.; Liu, R.; Gao, J.; Liu, M.D.; Huang, L.; Wang, G.Z.; Wang, D.; et al. Research advances in the structures and biological activities of secondary metabolites from Talaromyces. Front. Microbiol. 2022, 13, 984801. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, M.; Uchida, R.; Ohte, S.; Miyachi, N.; Kobayashi, K.; Sato, N.; Nonaka, K.; Masuma, R.; Fukuda, T.; Yasuhara, T.; et al. New dinapinone derivatives, potent inhibitors of triacylglycerol synthesis in mammalian cells, produced by Talaromyces pinophilus FKI-3864. J. Antibiot. 2013, 66, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Kuppers, L.; Ebrahim, W.; El-Neketi, M.; Ozkaya, F.C.; Mandi, A.; Kurtan, T.; Orfali, R.S.; Muller, W.E.G.; Hartmann, R.; Lin, W.; et al. Lactones from the sponge-derived fungus Talaromyces rugulosus. Mar. Drugs 2017, 15, 359. [Google Scholar] [CrossRef] [PubMed]
- Bara, R.; Zerfass, I.; Aly, A.H.; Goldbach-Gecke, H.; Raghavan, V.; Sass, P.; Mandi, A.; Wray, V.; Polavarapu, P.L.; Pretsch, A.; et al. Atropisomeric dihydroanthracenones as inhibitors of multiresistant Staphylococcus aureus. J. Med. Chem. 2013, 56, 3257–3272. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Sun, W.; Liu, X.; Wei, M.; Liang, Y.; Wang, J.; Zhu, H.; Zhang, Y. Anti-inflammatory spiroaxane and drimane sesquiterpenoids from Talaromyces minioluteus (Penicillium minioluteum). Bioorganic Chem. 2019, 91, 103166. [Google Scholar] [CrossRef] [PubMed]
- Hong, X.; Guan, X.; Lai, Q.; Yu, D.; Chen, Z.; Fu, X.; Zhang, B.; Chen, C.; Shao, Z.; Xia, J.; et al. Characterization of a bioactive meroterpenoid isolated from the marine-derived fungus Talaromyces sp. Appl. Microbiol. Biotechnol. 2022, 106, 2927–2935. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Oka, Y.; Kai, K.; Akiyama, K. New chrodrimanin congeners, chrodrimanins D-H, from YO-2 of Talaromyces sp. Biosci. Biotechnol. Biochem. 2012, 76, 1765–1768. [Google Scholar] [CrossRef] [PubMed]
- Schlingmann, G.; Milne, L.; Carter, G.T. Isolation and identification of antifungal polyesters from the marine fungus Hypoxylon oceanicum LL-15G256. Tetrahedron 2002, 58, 6825–6835. [Google Scholar] [CrossRef]
- Komai, S.I.; Hosoe, T.; Itabashi, T.; Nozawa, K.; Yaguchi, T.; Fukushima, K.; Kawai, K.I. New penicillide derivatives isolated from Penicillium simplicissimum. J. Nat. Med. 2006, 60, 185–190. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Yang, J.; Liao, Y.Y.; Cheng, G.; Chen, J.; Mo, S.; Yuan, L.; Cheng, X.D.; Qin, J.J.; Shao, Z. Aspeterreurone A, a cytotoxic dihydrobenzofuran-phenyl acrylate hybrid from the deep-sea-derived fungus Aspergillus terreus CC-S06-18. J. Nat. Prod. 2020, 83, 1998–2003. [Google Scholar] [CrossRef] [PubMed]
- Pracht, P.; Bohle, F.; Grimme, S. Automated exploration of the low-energy chemical space with fast quantum chemical methods. Phys. Chem. Chem. Phys. PCCP 2020, 22, 7169–7192. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Petersson, G.A.; Nakatsuji, H.; et al. Gaussian 16 Rev. C.01; Gaussian, Inc.: Wallingford, CT, USA, 2016. [Google Scholar]
- Grimblat, N.; Zanardi, M.M.; Sarotti, A.M. Beyond DP4: An improved probability for the stereochemical assignment of isomeric compounds using quantum chemical calculations of NMR shifts. J. Org. Chem. 2015, 80, 12526–12534. [Google Scholar] [CrossRef] [PubMed]
- Pescitelli, G.; Bruhn, T. Good computational practice in the assignment of absolute configurations by TDDFT calculations of ECD spectra. Chirality 2016, 28, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.; Chen, F. Multiwfn: A multifunctional wavefunction analyzer. J. Comput. Chem. 2012, 33, 580–592. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cheng, J.W.; Qin, J.J.; Hu, B.; Li, X.; Nijampatnam, B.; Velu, S.E.; Fan, J.; Yang, X.R.; Zhang, R. MDM2-NFAT1 dual inhibitor, MA242: Effective against hepatocellular carcinoma, independent of p53. Cancer Lett. 2019, 459, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liao, Y.; Zhang, B.; Gao, M.; Ke, W.; Li, F.; Shao, Z. Citrinin monomer and dimer derivatives with antibacterial and cytotoxic activities isolated from the deep sea-derived fungus Penicillium citrinum NLG-S01-P1. Mar. Drugs 2019, 17, 46. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Liao, Y.; Chen, R.; Hou, Y.; Ke, W.; Zhang, B.; Gao, M.; Shao, Z.; Chen, J.; Li, F. Chlorinated azaphilone pigments with antimicrobial and cytotoxic activities isolated from the deep sea derived fungus Chaetomium sp. NA-S01-R1. Mar. Drugs 2018, 16, 61. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xu, L.; Huang, W.; Li, F. Highly lethal Vibrio parahaemolyticus strains cause acute mortality in Penaeus vannamei post-larvae. Aquaculture 2022, 548, 737605. [Google Scholar] [CrossRef]







| 1 | 2 | 3 | ||||
|---|---|---|---|---|---|---|
| Position | δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) | δC, Type | δH, Mult. (J in Hz) |
| 1-β | 36.9, CH2 | 1.80 (dd, 11.7, 4.5 Hz, 1H) | 41.0, CH2 | 1.88 (dd, 12.1, 4.4 Hz, 1H) | 40.8, CH2 | 1.82 (dd, 11.9, 4.6 Hz, 1H) |
| 1-α | 36.9, CH2 | 1.58 (overlapped a, 1H) | 41.0, CH2 | 1.39 (t, 12.1 Hz, 1H) | 40.8, CH2 | 1.42 (overlapped a, 1H) |
| 2-β | 70.9, CH | 5.25 (ddd, 12.1, 4.4, 2.5 Hz, 1H) | 65.7, CH | 4.23 (ddd, 12.0, 4.4, 2.9 Hz, 1H) | 66.3, CH | 4.08 (dt, 11.9, 4.1 Hz, 1H) |
| 3-β | 76.5, CH | 3.53 (brs, 1H) | 80.3, CH | 4.93 (d, 2.8 Hz, 1H) | 78.7, CH | 3.48 (d, 2.9 Hz, 1H) |
| 4 | 38.5, C | - | 38.1, C | - | 37.9, C | - |
| 5 | 47.9, CH | 1.64 (d, 12.6, 1H) | 49.5, CH | 1.47 (overlapped a, 1H) | 47.9, CH | 1.53 (overlapped a, 1H) |
| 6-α | 18.9, CH2 | 1.72 (overlapped a, 1H) | 18.9, CH2 | 1.72 (overlapped a, 1H) | 19.0, CH2 | 1.72 (overlapped a, 1H) |
| 6-β | 18.9, CH2 | 1.40 (dd, 12.8, 3.4, 1H) | 18.9, CH2 | 1.42 (dd, 13.4, 3.4, 1H) | 19.0, CH2 | 1.40 (overlapped a, 1H) |
| 7-β | 40.4, CH2 | 2.09 (overlapped a, 1H) | 40.5, CH2 | 2.14 (dt, 13.6, 3.5 Hz, 1H) | 40.6, CH2 | 2.11 (dt, 11.2, 3.0 Hz, 1H) |
| 7-α | 40.4, CH2 | 1.72 (overlapped a, 1H) | 40.5, CH2 | 1.75 (overlapped a, 1H) | 40.6, CH2 | 1.71 (overlapped a, 1H) |
| 8 | 77.9, C | - | 77.7, C | - | 77.8, C | - |
| 9 | 51.2, CH | 1.76 (dd, 13.1, 5.1 Hz, 1H) | 51.6, CH | 1.78 (dd, 13.1, 5.0 Hz, 1H) | 51.5, CH | 1.75 (m, 1H) |
| 10 | 38.0, C | - | 37.8, C | - | 38.3, C | - |
| 11-α | 19.4, CH2 | 2.48 (dd, 15.9, 5.1 Hz, 1H) | 19.3, CH2 | 2.60 (m, 1H) | 19.4, CH2 | 2.59 (m, 1H) |
| 11-β | 19.4, CH2 | 2.25 (dd, 15.8, 13.1 Hz, 1H) | 19.3, CH2 | 2.29 (dd, 15.8, 13.2 Hz, 1H) | 19.4, CH2 | 2.28 (dd, 15.8, 13.1 Hz, 1H) |
| 12 | 21.0, CH3 | 1.17 (s, 3H) | 21.0, CH3 | 1.20 (s, 3H) | 20.9, CH3 | 1.18 (s, 3H) |
| 13 | 28.4, CH3 | 1.07 (s, 3H) | 28.1, CH3 | 0.95 (s, 3H) | 28.5, CH3 | 1.07 (s, 3H) |
| 14 | 21.7, CH3 | 0.95 (s, 3H) | 21.7, CH3 | 1.00 (s, 3H) | 21.6, CH3 | 0.91 (s, 3H) |
| 15 | 15.8, CH3 | 1.01 (s, 3H) | 16.0, CH3 | 0.99 (s, 3H) | 15.9, CH3 | 0.96 (s, 3H) |
| 16 | 170.4, C | - | 172.1, C | - | ||
| 17 | 21.4, CH3 | 2.11 (s, 3H) | 21.0, CH3 | 2.11 (s, 3H) | ||
| 1′ | 110.7, C | - | 110.2, C | - | 110.3, C | - |
| 2′ | 139.1, C | - | 141.8, C | - | 141.8, C | - |
| 3′ | 101.7, C | - | 106.6, C | - | 106.5, C | - |
| 4′ | 162.2, C | - | 161.5, C | - | 161.5, C | - |
| 5′ | 103.4, CH | 6.28 (s, 1H) | 99.7, CH | 6.33 (s, 1H) | 99.8, CH | 6.33 (s, 1H) |
| 6′ | 160.4, C | - | 158.5, C | - | 158.6, C | - |
| 7′a | 31.4, CH2 | 2.95 (dd, 16.7, 3.4 Hz, 1H) | 32.3, CH2 | 2.87 (dd, 16.4, 2.8 Hz, 1H) | 32.4, CH2 | 2.88 (dd, 16.4, 2.8 Hz, 1H) |
| 7′b | 31.4, CH2 | 2.61 (dd, 16.8, 11.5 Hz, 1H) | 32.3, CH2 | 2.57 (dd, 14.4, 9.5 Hz, 1H) | 32.4, CH2 | 2.57 (m, 1H) |
| 8′ | 74.7, CH | 4.63 (dqd, 12.8, 6.3, 3.0 Hz, 1H) | 72.8, CH | 4.45 (ddd, 9.0, 6.1, 2.7 Hz, 1H) | 72.8, CH | 4.48 (dtt, 12.7, 6.5, 3.2 Hz, 1H) |
| 9′ | 20.9, CH3 | 1.55 (d, 6.2 Hz, 3H) | 20.8, CH3 | 1.49 (d, 6.1 Hz, 3H) | 20.8, CH3 | 1.49 (d, 6.3 Hz, 3H) |
| 10′ | 170.1, C | - | 163.1, C | - | 163.2, C | - |
| 11′ | 56.1, CH3 | 3.86 (s, 3H) | 56.0, CH3 | 3.86 (s, 3H) | ||
| OH-4′ | - | 11.09 (s, 1H) | ||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, L.; Xia, J.; Chen, Z.; Lin, F.; Shao, Z.; Wang, W.; Hong, X. Cytotoxic and Antibacterial Meroterpenoids Isolated from the Marine-Derived Fungus Talaromyces sp. M27416. Mar. Drugs 2024, 22, 186. https://doi.org/10.3390/md22040186
Tang L, Xia J, Chen Z, Lin F, Shao Z, Wang W, Hong X. Cytotoxic and Antibacterial Meroterpenoids Isolated from the Marine-Derived Fungus Talaromyces sp. M27416. Marine Drugs. 2024; 22(4):186. https://doi.org/10.3390/md22040186
Chicago/Turabian StyleTang, Lingzhi, Jinmei Xia, Zhongwei Chen, Fengjiao Lin, Zongze Shao, Weiyi Wang, and Xuan Hong. 2024. "Cytotoxic and Antibacterial Meroterpenoids Isolated from the Marine-Derived Fungus Talaromyces sp. M27416" Marine Drugs 22, no. 4: 186. https://doi.org/10.3390/md22040186