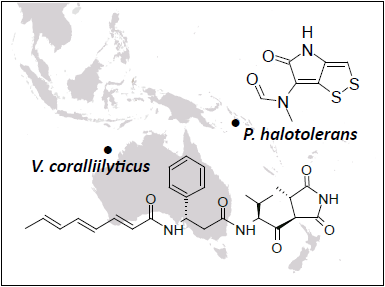
Antibacterial Compounds from Marine Vibrionaceae Isolated on a Global Expedition
Abstract
:1. Introduction
2. Results and Discussion
2.1. Selection of Strains with Pronounced Antibacterial Activity
2.2. Phylogenetic Identification and Chemotyping of Strains
2.3. Bioassay-Guided Identification of Antibacterial Compounds
3. Experimental Section
3.1. Isolation of Bioactive Marine Vibrionaceae
3.2. Selection of Strains with Pronounced Antibacterial Activity
3.3. Phylogenetic Analysis
3.4. Influence of Culture Conditions on Bioactivity
3.5. Chemotyping
3.6. Isolation and Structural Elucidation of Antibacterial Compounds
Andrimid
Holomycin
4. Conclusions
Acknowledgements
- Samples Availability: Available from the authors.
References
- Hibbing, ME; Fuqua, C; Parsek, MR; Peterson, SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol 2010, 8, 15–25. [Google Scholar]
- Demain, AL; Sanchez, S. Microbial drug discovery: 80 years of progress. J Antibiot 2009, 62, 5–16. [Google Scholar]
- Fischbach, MA. Antibiotics from microbes: Converging to kill. Curr Opin Microbiol 2009, 12, 520–527. [Google Scholar]
- Berdy, J. Bioactive Microbial Metabolites. J Antibiot 2005, 58, 1–26. [Google Scholar]
- Nair, S; Simidu, U. Distribution and Significance of Heterotrophic Marine Bacteria with Antibacterial Activity. Appl Environ Microbiol 1987, 53, 2957–2962. [Google Scholar]
- Long, E; Azam, F. Antagonistic Interactions among Marine Pelagic Bacteria. Appl Environ Microbiol 2001, 67, 4975–4983. [Google Scholar]
- Zhang, LX; An, R; Wang, JP; Sun, N; Zhang, S; Hu, JC; Kuai, J. Exploring novel bioactive compounds from marine microbes. Curr Opin Microbiol 2005, 8, 276–281. [Google Scholar]
- Gram, L; Melchiorsen, J; Bruhn, JB. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms.
- Hohmann, C; Schneider, K; Bruntner, C; Irran, E; Nicholson, G; Bull, AT; Jones, AL; Brown, R; Stach, JEM; Goodfellow, M; et al. Caboxamycin, a new antibiotic of the benzoxazole family produced by the deep-sea strain Streptomyces sp. 2009, 99–104. [Google Scholar]
- Taylor, MW; Radax, R; Steger, D; Wagner, M. Sponge-Associated Microorganisms: Evolution, Ecology, and Biotechnological Potential. Microbiol Mol Biol Rev 2007, 71, 295–347. [Google Scholar]
- Wiese, J; Thiel, V; Nagel, K; Staufenberger, T; Imhoff, JF. Diversity of Antibiotic-Active Bacteria Associated with the Brown Alga Laminaria saccharina from the Baltic Sea. Mar Biotechnol 2009, 11, 287–300. [Google Scholar]
- Rypien, KL; Ward, JR; Azam, F. Antagonistic interactions among coral-associated bacteria. Environ Microbiol 2010, 12, 28–39. [Google Scholar]
- Herndl, H; Wiese, J; Thiel, V; Imhoff, JM. Phylogenetic diversity and antimicrobial activities of bryozoan-associated bacteria isolated from Mediterranean and Baltic Sea habitats. Syst Appl Microbiol 2010, 33, 94–104. [Google Scholar]
- Bowman, JP. Bioactive Compound Synthetic Capacity and Ecological Significance of Marine Bacterial Genus Pseudoalteromonas. Mar Drugs 2007, 5, 220–241. [Google Scholar]
- Martens, T; Gram, L; Grossart, HP; Kessler, D; Muller, R; Simon, M; Wenzel, SC; Brinkhoff, T. Bacteria of the Roseobacter clade show potential for secondary metabolite production. Microb Ecol 2007, 54, 31–42. [Google Scholar]
- Bull, AT; Stach, JEM. Marine actinobacteria: new opportunities for natural product search and discovery. Trends Microbiol 2007, 15, 491–499. [Google Scholar]
- Andersen, RJ; Wolfe, MS; Faulkner, DJ. Autotoxic Antibiotic Production by a Marine Chromobacterium. Mar Biol 1974, 27, 281–285. [Google Scholar]
- Geng, HF; Bruhn, JB; Nielsen, KF; Gram, L; Belas, R. Genetic dissection of tropodithietic acid biosynthesis by marine roseobacters. Appl Environ Microbiol 2008, 74, 1535–1545. [Google Scholar]
- Romanenko, LA; Uchino, M; Kalinovskaya, NI; Mikhailov, VV. Isolation, phylogenetic analysis and screening of marine mollusc-associated bacteria for antimicrobial, hemolytic and surface activities. Microbiol Res 2008, 163, 633–644. [Google Scholar]
- Das, P; Mukherjee, S; Sen, R. Antimicrobial potential of a lipopeptide biosurfactant derived from a marine Bacillus circulans. J Appl Microbiol 2008, 104, 1675–1684. [Google Scholar]
- Kiran, GS; Thomas, TA; Selvin, J. Production of a new glycolipid biosurfactant from marine Nocardiopsis lucentensis MSA04 in solid-state cultivation. Colloids Surf B Biointerfaces 2010, 78, 8–16. [Google Scholar]
- Gomez, D; Espinosa, E; Bertazzo, M; Lucas-Elio, P; Solano, F; Sanchez-Amat, A. The macromolecule with antimicrobial activity synthesized by Pseudoalteromonas luteoviolacea strains is an l-amino acid oxidase. Appl Microbiol Biotechnol 2008, 79, 925–930. [Google Scholar]
- Thompson, FL; Iida, T; Swings, J. Biodiversity of Vibrios. Microbiol Mol Biol Rev 2004, 68, 403–431. [Google Scholar]
- Farmer, J; Hickman-Brenner, F. The Genera Vibrio and Photobacterium. In The Prokaryotes; Dworkin, M, Falkow, S, Rosenberg, E, Schleifer, KK-H, Stackebrandt, E, Eds.; Springer: New York, NY, USA, 2006. [Google Scholar]
- Austin, B; Stuckey, LF; Robertson, PAW; Effendi, I; Griffith, DRW. A Probiotic Strain of Vibrio alginolyticus Effective in Reducing Diseases Caused by Aeromonas salmonicida, Vibrio anguillarum and Vibrio ordalii. J Fish Dis 1995, 18, 93–96. [Google Scholar]
- Radjasa, OK; Sabdono, A; Zocchi, J; Zocchi, E. Richness of secondary metabolite-producing marine bacteria associated with the sponge Haliclona sp. Int J Pharmacol 2007, 3, 275–279. [Google Scholar]
- Hjelm, M; Riaza, A; Formoso, F; Melchiorsen, J; Gram, L. Seasonal Incidence of Autochthonous Antagonistic Roseobacter spp. and Vibrionaceae Strains in a Turbot Larva (Scophthalmus maximus) Rearing System. Appl Environ Microbiol 2004, 70, 7288–7294. [Google Scholar]
- Castro, D; Pujalte, MJ; Lopez-Cortes, L; Garay, E; Borrego, JJ. Vibrios isolated from the cultured manila clam (Ruditapes philippinarum): Numerical taxonomy and antibacterial activities. J Appl Microbiol 2002, 93, 438–447. [Google Scholar]
- Long, RA; Rowley, DC; Zamora, E; Liu, J; Bartlett, DH; Azam, F. Antagonistic Interactions among Marine Bacteria Impede the Proliferation of Vibrio cholerae. Appl Environ Microbiol 2005, 71, 8531–8536. [Google Scholar]
- Kobayashi, M; Aoki, S; Gato, K; Matsunami, K; Kurosu, M; Kitagawa, I. Marine Natural Products. XXXIV. Trisindoline, a New Antibiotic Indole Trimer, Produced by a Bacterium of Vibrio sp. Separated from the Marine Sponge Hyrtios altum. Chem Pharm Bull 1994, 42, 2449–2451. [Google Scholar]
- Oclarit, JM; Okada, H; Ohta, S; Kaminura, K; Yamaoka, Y; Iizuka, T; Miyashiro, S; Ikegami, S. Anti-Bacillus Substance in the Marine Sponge, Hyatella Species, Produced by an Associated Vibrio Species Bacterium. Microbios 1994, 78, 7–16. [Google Scholar]
- Vynne, NG; Mansson, M; Nielsen, KF; Gram, L. Bioactivity, chemical profiling and 16S rRNA based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar Biotechnol 2010. submitted for publication. [Google Scholar]
- Gram, L; Porsby, CH; Heilmann, J; Jensen, M; Melchiorsen, J; Nielsen, KF. A cosmopolitan bacterium: Phylogentic and phenotypic homogeneity in a global collection of Ruegeria mobilis of the Roseobacter clade. Appl Environ Microbiol 2010. submitted for publication. [Google Scholar]
- Thompson, FL; Gevers, D; Thompson, CC; Dawyndt, P; Naser, S; Hoste, B; Munn, CB; Swings, J. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl Environ Microbiol 2005, 71, 5107–5115. [Google Scholar]
- Pascual, J; Macian, MC; Arahal, DR; Garay, E; Pujalte, MJ. Multilocus sequence analysis of the central clade of the genus Vibrio by using 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int J Syst Evol Microbiol 2009, 60, 154–165. [Google Scholar]
- Altschul, SF; Madden, TL; Schaffer, AA; Zhang, JH; Zhang, Z; Miller, W; Lipman, DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res 1997, 25, 3389–3402. [Google Scholar]
- Nielsen, KF; Smedsgaard, J. Fungal metabolite screening: database of 474 mycotoxins and fungal metabolites for dereplication by standardised liquid chromatography-UV-mass spectrometry methodology. J Chromatogr A 2003, 1002, 111–136. [Google Scholar]
- Mitova, M; Popov, S; De Rosa, S. Cyclic peptides from a Ruegeria strain of bacteria associated with the sponge Suberites domuncula. J Nat Prod 2004, 67, 1178–1181. [Google Scholar]
- Rungprom, W; Siwu, ERO; Lambert, LK; Dechsakulwatana, C; Barden, MC; Kokpol, U; Blanchfield, JT; Kita, M; Garson, MJ. Cyclic tetrapeptides from marine bacteria associated with the seaweed Diginea sp. and the sponge Halisarca ectofibrosa. Tetrahedron 2008, 64, 3147–3152. [Google Scholar]
- Mansson, M; Nielsen, A; Kjaerulff, L; Gotfredsen, CH; Ingmer, H; Wietz, M; Gram, L; Larsen, TO. Inhibition of virulence gene expression in Staphylococcus aureus by novel depsipeptides from a marine. Photobacterium 2010. to be submitted for publication. [Google Scholar]
- Chien, CC; Chen, CC; Choi, MH; Kung, SS; Wei, YH. Production of poly-β-hydroxybutyrate (PHB) by Vibrio spp. isolated from marine environment. J Biotechnol 2007, 132, 259–263. [Google Scholar]
- Bull, AT. Microbial Diversity and Bioprospecting; ASM Press: Washington, DC, USA, 2003. [Google Scholar]
- Dieckmann, R; Strauch, E; Alter, T. Rapid identification and characterization of Vibrio species using whole-cell MALDI-TOF mass spectrometry. J Appl Microbiol 2010, 109, 199–211. [Google Scholar]
- Jensen, PR; Williams, PG; Oh, DC; Zeigler, L; Fenical, W. Species-Specific Secondary Metabolite Production in Marine Actinomycetes of the Genus Salinispora. Appl Environ Microbiol 2007, 73, 1146–1152. [Google Scholar]
- Mansson, M; Phipps, RK; Gram, L; Munro, MHG; Larsen, TO; Nielsen, KF. Explorative Solid-Phase Extraction (E-SPE) for Accelerated Microbial Natural Product Discovery, Dereplication, and Purification. J Nat Prod 2010, 73, 1126–1132. [Google Scholar]
- Fredenhagen, A; Tamura, SY; Kenny, PTM; Komura, H; Naya, Y; Nakanishi, K; Nishiyama, K; Sugiura, M; Kita, H. Andrimid, a new peptide antibiotic produced by an intracellular bacterial symbiont isolated from a brown planthopper. J Am Chem Soc 1987, 109, 4409–4411. [Google Scholar]
- Needham, J; Kelly, MT; Ishige, M; Andersen, RJ. Andrimid and moiramides A–C, metabolites produced in culture by a marine isolate of the bacterium Pseudomonas fluorescens: Structure elucidation and biosynthesis. J Org Chem 1994, 59, 2058–2063. [Google Scholar]
- Jin, M; Fischbach, MA; Clardy, J. A Biosynthetic Gene Cluster for the Acetyl-CoA Carboxylase Inhibitor Andrimid. J Am Chem Soc 2006, 128, 10660–10661. [Google Scholar]
- Freiberg, C; Brunner, NA; Schiffer, G; Lampe, T; Pohlmann, J; Brands, M; Raabe, M; Häbich, D; Ziegelbauer, K. Identification and Characterization of the First Class of Potent Bacterial Acetyl-CoA Carboxylase Inhibitors with Antibacterial Activity. J Biol Chem 2004, 279, 26066–26073. [Google Scholar]
- Singh, MP; Mroczenski-Wildey, MJ; Steinberg, DA; Andersen, RJ; Maiese, WM; Greenstein, M. Biological activity and mechanistic studies of andrimid. J Antibiot 1997, 50, 270–273. [Google Scholar]
- Fischbach, MA; Walsh, CT; Clardy, J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc Natl Acad Sci USA 2008, 105, 4601–4608. [Google Scholar]
- Oliva, B; O’Neill, A; Wilson, JM; O’Hanlon, PJ; Chopra, I. Antimicrobial Properties and Mode of Action of the Pyrrothine Holomycin. Antimicrob Agents Chemother 2001, 45, 532–539. [Google Scholar]
- Shiozawa, H; Kagasaki, T; Kinoshita, T; Haruyama, H; Domon, H; Utsui, Y; Kodama, K; Takahashi, S. Thiomarinol, A New Hybrid Antimicrobial Antibiotic Produced by A Marine Bacterium. Fermentation, Isolation, Structure, and Antimicrobial Activity. J Antibiot 1993, 46, 1834–1842. [Google Scholar]
- Kenig, M; Reading, C. Holomycin and an Antibiotic (Mm-19290) Related to Tunicamycin, Metabolites of Streptomyces clavuligerus. J Antibiot 1979, 32, 549–554. [Google Scholar]
- Hou, YH; Li, FC; Wang, SJ; Qin, S; Wang, QF. Intergeneric conjugation in holomycin-producing marine Streptomyces sp. strain M095. Microbiol Res 2008, 163, 96–104. [Google Scholar]
- Ben-Haim, Y; Thompson, FL; Thompson, CC; Cnockaert, MC; Hoste, B; Swings, J; Rosenberg, E. Vibrio coralliilyticus sp. nov., a temperature-dependent pathogen of the coral Pocillopora damicornis. Int J Syst Evol Microbiol 2003, 53, 309–315. [Google Scholar]
- Prado, S; Romalde, JL; Montes, J; Barja, JL. Pathogenic bacteria isolated from disease outbreaks in shellfish hatcheries. First description of Vibrio neptunius as an oyster pathogen. Dis Aquat Org 2005, 67, 209–215. [Google Scholar]
- Li, JW-H; Vederas, JC. Drug discovery and natural products: End of an era or an endless frontier. Science 2009, 325, 161–165. [Google Scholar]
- Laatsch, H. Antibase; Wiley-VCH: Weinheim, Germany, 2010. Available online: http://www.users.gwdg.de/~ucoc/laatschAntibase.htm accessed on 1 November 2010.
- Lang, G; Mayhudin, NA; Mitova, MI; Sun, L; van der Sar, S; Blunt, JW; Cole, ALJ; Ellis, G; Laatsch, H; Munro, MHG. Evolving trends in the dereplication of natural product extracts: New methodology for rapid, small-scale investigation of natural product extracts. J Nat Prod 2008, 71, 1595–1599. [Google Scholar]
- Kind, T; Fiehn, O. Metabolomic database annotations via query of elemental compositions: Mass accuracy is insufficient even at less than 1 ppm. BMC Bioinformatics 2006, 7, 234. [Google Scholar]
- Hjelm, M; Bergh, I; Riaza, A; Nielsen, J; Melchiorsen, J; Jensen, S; Duncan, H; Ahrens, P; Birkbeck, H; Gram, L. Selection and Identification of Autochthonous Potential Probiotic Bacteria from Turbot Larvae (Scophthalmus maximus) Rearing Units. Syst Appl Microbiol 2004, 27, 360–371. [Google Scholar]
- Ostling, J; Goodman, A; Kjelleberg, S. Behaviour of IncP-1 plasmids and a miniMu transposon in a marine Vibrio sp.: Isolation of starvation inducible lac operon fusions. FEMS Microbiol Lett 1991, 86, 83–93. [Google Scholar]



| Inhibition of | |||
|---|---|---|---|
| Strain | Species | V. anguillarum | S. aureus |
| S2052 | V. coralliilyticus | +++ | ++ |
| S2394 | V. neptunius | ++ | − |
| S2753 | P. halotolerans | +++ | ++ |
| S4051 | V. neptunius | ++ | − |
| S4053 | V. coralliilyticus | ++ | + |
© 2010 by the authors, licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wietz, M.; Mansson, M.; Gotfredsen, C.H.; Larsen, T.O.; Gram, L. Antibacterial Compounds from Marine Vibrionaceae Isolated on a Global Expedition. Mar. Drugs 2010, 8, 2946-2960. https://doi.org/10.3390/md8122946
Wietz M, Mansson M, Gotfredsen CH, Larsen TO, Gram L. Antibacterial Compounds from Marine Vibrionaceae Isolated on a Global Expedition. Marine Drugs. 2010; 8(12):2946-2960. https://doi.org/10.3390/md8122946
Chicago/Turabian StyleWietz, Matthias, Maria Mansson, Charlotte H. Gotfredsen, Thomas O. Larsen, and Lone Gram. 2010. "Antibacterial Compounds from Marine Vibrionaceae Isolated on a Global Expedition" Marine Drugs 8, no. 12: 2946-2960. https://doi.org/10.3390/md8122946