Studies on Synthesis and Structure-Activity Relationship (SAR) of Derivatives of a New Natural Product from Marine Fungi as Inhibitors of Influenza Virus Neuraminidase
Abstract
:1. Introduction
2. Results and Discussion
2.1. Chemistry
2.2. H1N1 Virus Neuraminidase Inhibition
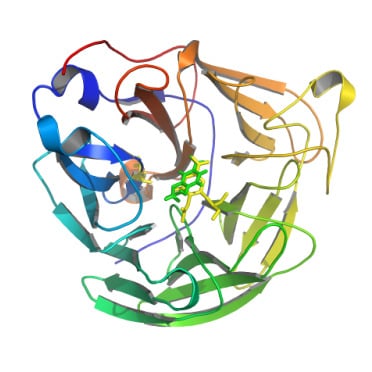
2.3. Molecular Docking Study
3. Experimental Section
3.1. Chemistry
3.2. Synthetic Methods of Compounds
3.2.1. General Procedure 1: Synthesis of Compounds 1–5, 7–13, 16–25
methyl 3-hydroxy-4-((3-methylbut-2-en-1-yl)oxy)benzoate (1)
methyl 3-hydroxy-4-(isopentyloxy)benzoate (2)
methyl 3-hydroxy-4-propoxybenzoate (3)
methyl 4-(allyloxy)-3-hydroxybenzoate (4)
methyl 3-hydroxy-4-(prop-2-yn-1-yloxy)benzoate (5)
methyl 3,4-bis(benzyloxy)benzoate (7)
methyl 3,4-dipropoxybenzoate (8)
3-hydroxy-4-((3-methylbut-2-en-1-yl)oxy)benzaldehyde (9)
3-hydroxy-4-(isopentyloxy)benzaldehyde (10)
3-hydroxy-4-propoxybenzaldehyde (11)
3-hydroxy-4-(prop-2-yn-1-yloxy)benzaldehyde (12)
4-(allyloxy)-3-hydroxybenzaldehyde (13)
3,4-bis((3-methylbut-2-en-1-yl)oxy)benzaldehyde (16)
3,4-bis(prop-2-yn-1-yloxy)benzaldehyde (17)
3,4-bis(allyloxy)benzaldehyde (18)
3,4-bis(isopentyloxy)benzaldehyde (19)
3,4-dipropoxybenzaldehyde (20)
3,4-bis(prop-2-yn-1-yloxy)benzoic acid (21)
3,4-bis(isopentyloxy)benzoic acid (22)
3,4-dipropoxybenzoic acid (23)
(E)-methyl 3-(3-hydroxy-4-(prop-2-yn-1-yloxy)phenyl)acrylate (24)
(E)-methyl 3-(3,4-bis(prop-2-yn-1-yloxy)phenyl)acrylate (25)
3.2.2. General Procedure 2: Synthesis of Compounds 6, 14–15
methyl 3-(allyloxy)-4-hydroxybenzoate (6)
4-hydroxy-3-((3-methylbut-2-en-1-yl)oxy)benzaldehyde (14)
3-(allyloxy)-4-hydroxybenzaldehyde (15)
(E)-methyl 3-(3,4-dihydroxyphenyl)acrylate (26)
methyl 3-(isopentyloxy)-4-methoxybenzoate (27)
3-methylbut-2-en-1-yl 4-methoxy-3-((3-methylbut-2-en-1-yl)oxy)benzoate (28)
3-methylbut-2-en-1-yl 3-hydroxy-4-methoxybenzoate (29)
3.2.3. Synthesis of 3,4-bis(prop-2-yn-1-yloxy)benzamide (30)
methyl 3-hydroxy-4-methoxybenzoate (31)
(E)-methyl 3-(4-hydroxy-3-((3-methylbut-2-en-1-yl)oxy)phenyl)acrylate (32)
3.3. Virus and Cell Culture
3.4. Neuraminidase Activity Inhibition Assay
3.5. Modelling of Enzyme-Substrate Complexes by Molecular Docking
4. Conclusions
Acknowledgments
- Samples Availability: Available from the authors
References
- Webster, RG; Bean, WJ; Gorman, OT; Chambers, TM; Kawaoka, Y. Evolution and ecology of influenza A viruses. Microbiol. Rev 1992, 56, 152–179. [Google Scholar]
- Cox, N; Subbarao, K. Global epidemiology of influenza: Past and present. Annu. Rev. Med 2000, 51, 407–421. [Google Scholar]
- Atigadda, VR; Brouillette, WJ; Duarte, F; Babu, YS; Bantia, S; Chand, P; Chu, N; Montgomery, JA; Walsh, DA; Sudbeck, E. Hydrophobic benzoic acids as inhibitors of influenza neuraminidase. Bioorg. Med. Chem 1999, 7, 2487–2497. [Google Scholar]
- Chand, P; Kotian, PL; Morris, PE; Bantia, S; Walsh, DA; Babu, YS. Synthesis and inhibitory activity of benzoic acid and pyridine derivatives on influenza neuraminidase. Bioorg. Med. Chem 2005, 13, 2665–2678. [Google Scholar]
- Magesh, S; Savita, V; Moriya, S; Suzuki, T; Miyagi, T; Ishida, H; Kiso, M. Human sialidase inhibitors: Design, synthesis, and biological evaluation of 4-acetamido-5-acylamido-2-fluoro benzoic acids. Bioorg. Med. Chem 2009, 17, 4595–4603. [Google Scholar]
- Jedrzejas, MJ; Singh, S; Brouillette, WJ; Laver, WG; Air, GM; Luo, M. Structure of aromatic inhibitors of influenza virus neuraminidase. Biochemistry 1995, 34, 3144–3151. [Google Scholar]
- Chand, P; Babu, YS; Bantia, S; Chu, N; Cole, LB; Kotian, PL; Laver, WG; Montgomery, JA; Pathak, VP; Petty, SL; Shrout, DP; Walsh, DA; Walsh, GM. Design and synthesis of benzoic acid derivatives as influenza neuraminidase inhibitors using structure-based drug design. J. Med. Chem 1997, 40, 4030–4052. [Google Scholar]
- Shao, C; Guo, Z; Peng, H; Peng, G; Huang, Z; She, Z; Lin, Y; Zhou, S. A new isoprenyl phenyl ether compound from mangrove fungus. Chem. Nat. Comp 2007, 43, 377–380. [Google Scholar]
- Wang, T; Wade, RC. Comparative binding energy (COMBINE) analysis of influenza neuraminidase-inhibitor complexes. J. Med. Chem 2001, 44, 961–971. [Google Scholar]
- Hayashida, M; Ishizaki, M; Hara, H. Investigation of Selective Mono-deallylation of O,O′-Diallylcatechols and 3-Methylene-1,5-benzodioxepanes. Chem. Pharm. Bull 2006, 54, 1299–1303. [Google Scholar]
- Kiso, M; Kubo, S; Ozawa, M; Quynh, ML; Nidom, CA; Yamashita, M; Kawaoka, Y. Efficacy of the New Neuraminidase Inhibitor CS-8958 against H5N1 Influenza Viruses. PLoS Pathog 2010, 6, e1000786. [Google Scholar]
- Bliss, C. The calculation of the dose-mortality curve. Ann. Appl. Biol 1935, 22, 134–167. [Google Scholar]
- Russell, RJ; Haire, LF; Stevens, DJ; Collins, PJ; Lin, YP; Blackburn, GM; Hay, AJ; Gamblin, SJ; Skehel, JJ. The structure of H5N1 avian influenza neuraminidase suggests new opportunities for drug design. Nature 2006, 443, 45–49, Samples Availability: Available from the authors.. [Google Scholar]








© 2011 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Li, J.; Zhang, D.; Zhu, X.; He, Z.; Liu, S.; Li, M.; Pang, J.; Lin, Y. Studies on Synthesis and Structure-Activity Relationship (SAR) of Derivatives of a New Natural Product from Marine Fungi as Inhibitors of Influenza Virus Neuraminidase. Mar. Drugs 2011, 9, 1887-1901. https://doi.org/10.3390/md9101887
Li J, Zhang D, Zhu X, He Z, Liu S, Li M, Pang J, Lin Y. Studies on Synthesis and Structure-Activity Relationship (SAR) of Derivatives of a New Natural Product from Marine Fungi as Inhibitors of Influenza Virus Neuraminidase. Marine Drugs. 2011; 9(10):1887-1901. https://doi.org/10.3390/md9101887
Chicago/Turabian StyleLi, Jing, Dingmei Zhang, Xun Zhu, Zhenjian He, Shu Liu, Mengfeng Li, Jiyan Pang, and Yongcheng Lin. 2011. "Studies on Synthesis and Structure-Activity Relationship (SAR) of Derivatives of a New Natural Product from Marine Fungi as Inhibitors of Influenza Virus Neuraminidase" Marine Drugs 9, no. 10: 1887-1901. https://doi.org/10.3390/md9101887
APA StyleLi, J., Zhang, D., Zhu, X., He, Z., Liu, S., Li, M., Pang, J., & Lin, Y. (2011). Studies on Synthesis and Structure-Activity Relationship (SAR) of Derivatives of a New Natural Product from Marine Fungi as Inhibitors of Influenza Virus Neuraminidase. Marine Drugs, 9(10), 1887-1901. https://doi.org/10.3390/md9101887