Bioactive Eunicellin-Based Diterpenoids from the Soft Coral Cladiella krempfi
Abstract
:1. Introduction
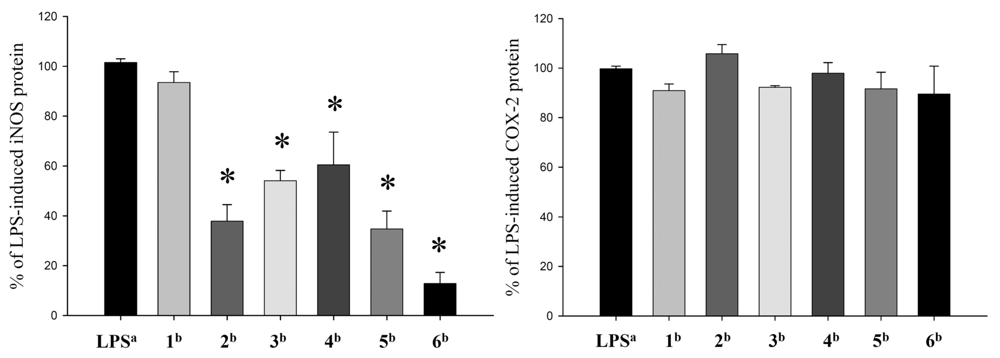
2. Results and Discussion
3. Experimental Section
3.1. General Experimental Procedures
3.2. Animal Material
3.3. Extraction and Separation
3.4. Cytotoxicity Testing
3.5. In Vitro Anti-Inflammatory Assay
Acknowledgements
- Samples Availability: Not available.
References
- Sarma, NS; Chavakula, R; Rao, IN; Kadirvelraj, R; Row, TNG; Saito, I. Crystal and molecular structure of sclerophytin F methyl ether from the soft coral Cladiella krempfi. J. Nat. Prod 1993, 56, 1977–1980. [Google Scholar]
- Lan, W-J; Lin, C-W; Su, J-Y; Zeng, L-M. Two steroidal glycosides from the soft coral Cladiella krempfi. Chem. J. Chin. Univ 2003, 24, 2019–2021. [Google Scholar]
- Huang, X-P; Deng, Z-W; Ofwegen, LV; Li, J; Fu, H-Z; Zhu, X-B; Lin, W-H. Two new pregnane glycosides from soft coral Cladiella krempfi. J. Asian Nat. Prod. Res 2006, 8, 287–291. [Google Scholar]
- Huang, X-P; Deng, Z-W; Zhu, X-B; Ofwegen, LV; Proksch, P; Lin, W-H. Krempenes A–D: A series of unprecedented pregnane-type steroids from the marine soft coral Cladiella krempfi. Helv. Chim. Acta 2006, 89, 2020–2026. [Google Scholar]
- Wang, G-H; Sheu, J-H; Chiang, MY; Lee, T-J. Pachyclavulariaenones A–C, three novel diterpenoids from the soft coral Pachyclavularia violacea. Tetrahedron Lett 2001, 42, 2333–2336. [Google Scholar]
- Wang, G-H; Sheu, J-H; Duh, C-Y; Chiang, MY. Pachyclavulariaenones D–G, new diterpenoids from the soft coral Pachyclavularia violacea. J. Nat. Prod 2002, 65, 1475–1478. [Google Scholar]
- Ahmed, AF; Wu, M-H; Wang, G-H; Wu, Y-C; Sheu, J-H. Eunicellin-based diterpenoids, australins A–D, isolated from the soft coral Cladiella australis. J. Nat. Prod 2005, 68, 1051–1055. [Google Scholar]
- Chen, B-W; Chang, S-M; Huang, C-Y; Chao, C-H; Su, J-H; Wen, Z-H; Hsu, C-H; Dai, C-F; Wu, Y-C; Sheu, J-H. Hirsutalins A–H, eunicellin-based diterpenoids from the soft coral Cladiella hirsuta. J. Nat. Prod 2010, 73, 1785–1791. [Google Scholar]
- Su, J-H; Huang, H-C; Chao, C-H; Yan, L-Y; Wu, Y-C; Wu, C-C; Sheu, J-H. Vigulariol, a new metabolite from the sea pen Vigularia juncea. Bull. Chem. Soc. Jpn 2005, 78, 877–879. [Google Scholar]
- Wu, S-L; Su, J-H; Wen, Z-H; Hsu, C-H; Chen, B-W; Dai, C-F; Kuo, Y-H; Sheu, J-H. Simplexins A–I, eunicellin-based diterpenoids from the soft coral Klyxum simplex. J. Nat. Prod 2009, 72, 994–1000. [Google Scholar]
- Wu, S-L; Su, J-H; Lu, Y; Chen, B-W; Huang, C-Y; Wen, Z-H; Kuo, Y-H; Sheu, J-H. Simplexins J–O, eunicellin-based diterpenoids from a Dongsha Atoll soft coral Klyxum simplex. Bull. Chem. Soc. Jpn 2011, 84, 626–632. [Google Scholar]
- Chen, B-W; Wu, Y-C; Chiang, MY; Su, J-H; Wang, W-H; Fan, T-Y; Sheu, J-H. Eunicellin-based diterpenoids from the cultured soft coral Klyxum simplex. Tetrahedron 2009, 65, 7016–7022. [Google Scholar]
- Chen, B-W; Chao, C-H; Su, J-H; Wen, Z-H; Sung, P-J; Sheu, J-H. Anti-inflammatory eunicellin-based diterpenoids from the cultured soft coral Klyxum simplex. Org. Biomol. Chem 2010, 8, 2363–2366. [Google Scholar]
- Chen, B-W; Chao, C-H; Su, J-H; Tsai, C-W; Wang, W-H; Wen, Z-H; Huang, C-Y; Sung, P-J; Wu, Y-C; Sheu, J-H. Klysimplexins I–T, eunicellin-based diterpenoids from the cultured soft coral Klyxum simplex. Org. Biomol. Chem 2011, 9, 834–844. [Google Scholar]
- Chen, Y-H; Tai, C-Y; Su, Y-D; Chang, Y-C; Lu, M-C; Weng, C-F; Su, J-H; Hwang, T-L; Wu, Y-C; Sung, P-J. Discovery of new eunicellins from an Indonesian octocoral Cladiella sp. Mar. Drugs 2011, 9, 934–943. [Google Scholar]
- Miyamoto, T; Yamada, K; Ikeda, N; Komori, T; Higuchi, R. Bioactive terpenoids from octocorallia, I. Bioactive diterpenoids: Litophynols A and B from the mucus of the soft coral Litophyton sp. J. Nat. Prod 1994, 57, 1212–1219. [Google Scholar]
- Rao, CB; Rao, DS; Satyanarayana, C; Rao, DV; Kassühlke, KE; Faulkner, DJ. New cladiellane diterpenes from the soft coral Cladiella australis of the Andaman and Nicobar Islands. J. Nat. Prod 1994, 57, 574–580. [Google Scholar]
- Alley, MC; Scudiero, DA; Monks, A; Hursey, ML; Czerwinski, MJ; Fine, DL; Abbott, BJ; Mayo, JG; Shoemaker, RH; Boyd, MR. Feasibility of drug screening with panels of human tumor cell lines using a microculture tetrazolium assay. Cancer Res 1988, 48, 589–601. [Google Scholar]
- Scudiero, DA; Shoemaker, RH; Paull, KD; Monks, A; Tierney, S; Nofziger, TH; Currens, MJ; Seniff, D; Boyd, MR. Evaluation of a soluble tetrazolium/formazan assay for cell growth and drug sensitivity in culture using human and other tumor cell lines. Cancer Res 1988, 48, 4827–4833. [Google Scholar]
- Jean, Y-H; Chen, W-F; Sung, C-S; Duh, C-Y; Huang, S-Y; Lin, C-S; Tai, M-H; Tzeng, S-F; Wen, Z-H. Capnellene, a natural marine compound derived from soft coral, attenuates chronic constriction injury-induced neuropathic pain in rats. Br. J. Pharmacol 2009, 158, 713–725. [Google Scholar]
- Jean, Y-H; Chen, W-F; Duh, C-Y; Huang, S-Y; Hsu, C-H; Lin, C-S; Sung, C-S; Chen, I-M; Wen, Z-H. Inducible nitric oxide synthase and cyclooxygenase-2 participate in anti-inflammatory and analgesic effects of the natural marine compound lemnalol from Formosan soft coral Lemnalia cervicorni. Eur. J. Pharmacol 2008, 578, 323–331. [Google Scholar]



| 1 a | 2 a | 3 b | 4 c | |
|---|---|---|---|---|
| 1 | 43.6 (CH) d | 45.5 (CH) | 45.5 (CH) | 43.9 (CH) |
| 2 | 90.5 (CH) | 92.3 (CH) | 92.6 (CH) | 92.6 (CH) |
| 3 | 86.0 (qC) | 86.1 (qC) | 86.1 (qC) | 85.9 (qC) |
| 4 | 34.0 (CH2) | 36.2 (CH2) | 35.4 (CH2) | 35.4 (CH2) |
| 5 | 30.0 (CH2) | 26.6 (CH2) | 28.5 (CH2) | 26.4 (CH2) |
| 6 | 77.0 (CH) | 88.3 (CH) | 82.2 (CH) | 87.4 (CH) |
| 7 | 79.3 (qC) | 78.7 (qC) | 78.2 (qC) | 78.4 (qC) |
| 8 | 78.8 (CH) | 79.6 (CH) | 80.1 (CH) | 79.1 (CH) |
| 9 | 81.9 (CH) | 81.7 (CH) | 81.4 (CH) | 82.5 (CH) |
| 10 | 50.3 (CH) | 53.2 (CH) | 53.5 (CH) | 50.7 (CH) |
| 11 | 143.7 (qC) | 148.4 (qC) | 148.6 (qC) | 143.2 (qC) |
| 12 | 72.9 (CH) | 31.7 (CH2) | 31.5 (CH2) | 73.1 (CH) |
| 13 | 29.2 (CH2) | 24.9 (CH2) | 24.9 (CH2) | 28.9 (CH2) |
| 14 | 37.9 (CH) | 44.2 (CH) | 44.1 (CH) | 37.6 (CH) |
| 15 | 23.2 (CH3) | 23.0 (CH3) | 22.8 (CH3) | 23.1 (CH3) |
| 16 | 17.8 (CH3) | 18.3 (CH3) | 18.4 (CH3) | 18.5 (CH3) |
| 17 | 114.9 (CH2) | 114.9 (CH2) | 110.6 (CH2) | 116.0 (CH2) |
| 18 | 28.9 (CH) | 29.0 (CH) | 29.0 (CH) | 28.8 (CH) |
| 19 | 21.5 (CH3) | 21.9 (CH3) | 21.9 (CH3) | 21.8 (CH3) |
| 20 | 16.6 (CH3) | 15.8 (CH3) | 15.5 (CH3) | 16.2 (CH3) |
| 3-n-butyrate | 172.3 (qC) | 172.3 (qC) | 172.4 (qC) | 172.3 (qC) |
| 37.3 (CH2) | 37.4 (CH2) | 37.4 (CH2) | 37.3 (CH2) | |
| 18.3 (CH2) | 18.3 (CH2) | 18.4 (CH2) | 18.3 (CH2) | |
| 13.6 (CH3) | 13.6 (CH3) | 13.6 (CH3) | 13.6 (CH3) | |
| 6-OMe | 56.9 (CH3) | 56.8 (CH3) | ||
| 6-OAc | 171.9 (qC) | |||
| 21.4 (CH3) | ||||
| 12-OAc | 170.5 (qC) | 170.3 (qC) | ||
| 21.9 (CH3) | 21.6 (CH3) |
| 1 a | 2 a | 3 b | 4 c | |
|---|---|---|---|---|
| 1 | 2.34 m | 2.25 m | 2.22 m | 2.32 m |
| 2 | 3.69 s | 3.60 s | 3.64 s | 3.64 s |
| 4 | 1.93 m; 2.43 m | 1.76 m; 2.66 dd (14.4, 9.0) | 1.97 m; 2.59 dd (15.2, 8.8) | 1.84 m; 2.55 dd (14.0, 10.0) |
| 5 | 1.50 m; 1.71 m | 1.36 m; 1.63 m | 1.46 m; 1.51 m | 1.35 m; 1.68 m |
| 6 | 4.63 d (7.2) d | 4.13 d (6.3) | 5.72 d (4.4) | 4.12 d (8.5) |
| 8 | 3.48 d (9.3) | 3.54 t (9.3) | 3.58 brt (9.2) | 3.45 t (9.5) |
| 9 | 4.05 dd (9.3, 5.7) | 3.89 dd (9.3, 6.9) | 3.84 dd (9.2, 7.2) | 4.09 dd (9.5, 6.5) |
| 10 | 3.30 t (5.7) | 3.32 t (6.9) | 3.36 t (7.2) | 3.33 t (6.5) |
| 12 | 5.44 d (3.3) | 2.06 m; 2.32 m | 2.05 m; 2.31 m | 5.46 dd (5.0, 2.5) |
| 13 | 1.50 m; 1.88 m | 1.07 m; 1.75 m | 1.06 m; 1.76 m | 1.46 m; 1.90 m |
| 14 | 1.68 m | 1.29 m | 1.25 m | 1.67 m |
| 15 | 1.47 s | 1.41 s | 1.38 s | 1.46 s |
| 16 | 1.26 s | 1.24 s | 1.29 s | 1.23 s |
| 17 | 5.20 brs | 4.79 s, 4.90 s | 4.79 s, 4.89 s | 5.20 s, 5.21 s |
| 18 | 1.85 m | 1.72 m | 1.69 m | 1.82 m |
| 19 | 0.97 d (6.6) | 0.96 d (6.6) | 0.97 d (6.8) | 0.96 d (6.5) |
| 20 | 0.86 d (6.6) | 0.79 d (6.6) | 0.79 d (6.8) | 0.85 d (6.5) |
| 3-n-butyrate | 2.28 m | 2.34 m | 2.31 m, 2.34 m | 2.17 m, 2.29 m |
| 1.65 m | 1.67 m | 1.67 m | 1.59 m | |
| 0.97 t (7.2) | 0.99 t (7.5) | 0.99 t (7.2) | 0.97 t (7.5) | |
| 6-OMe | 3.36 s | 3.36 s | ||
| 6-OAc | 2.09 s | |||
| 12-OAc | 2.09 s | 2.08 s |
© 2011 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Tai, C.-J.; Su, J.-H.; Huang, M.-S.; Wen, Z.-H.; Dai, C.-F.; Sheu, J.-H. Bioactive Eunicellin-Based Diterpenoids from the Soft Coral Cladiella krempfi. Mar. Drugs 2011, 9, 2036-2045. https://doi.org/10.3390/md9102036
Tai C-J, Su J-H, Huang M-S, Wen Z-H, Dai C-F, Sheu J-H. Bioactive Eunicellin-Based Diterpenoids from the Soft Coral Cladiella krempfi. Marine Drugs. 2011; 9(10):2036-2045. https://doi.org/10.3390/md9102036
Chicago/Turabian StyleTai, Chi-Jen, Jui-Hsin Su, Ming-Shyan Huang, Zhi-Hong Wen, Chang-Feng Dai, and Jyh-Horng Sheu. 2011. "Bioactive Eunicellin-Based Diterpenoids from the Soft Coral Cladiella krempfi" Marine Drugs 9, no. 10: 2036-2045. https://doi.org/10.3390/md9102036
APA StyleTai, C.-J., Su, J.-H., Huang, M.-S., Wen, Z.-H., Dai, C.-F., & Sheu, J.-H. (2011). Bioactive Eunicellin-Based Diterpenoids from the Soft Coral Cladiella krempfi. Marine Drugs, 9(10), 2036-2045. https://doi.org/10.3390/md9102036





