Production of Bioactive Secondary Metabolites by Marine Vibrionaceae
Abstract
:1. Introduction
1.1. Occurrence and Ecological Significance
1.2. Genomic Diversity and Phylogeny
2. Natural Product Production by Members of the Vibrionaceae Family
2.1. Compounds with Antibacterial Activity
2.2. Siderophores
2.3. Compounds with Other Activities
2.4. Compounds with Unknown Activities
3. Conclusion
Acknowledgments
References and Notes
- Thompson, FL; Iida, T; Swings, J. Biodiversity of vibrios. Microbiol. Mol. Biol. Rev 2004, 68, 403–431. [Google Scholar]
- Colwell, R; Swings, J; Thompson, FL. Association of Vibrio Biologists (AViB). Available online: http://www.vibriobiology.net/ accessed on 16 August 2011.
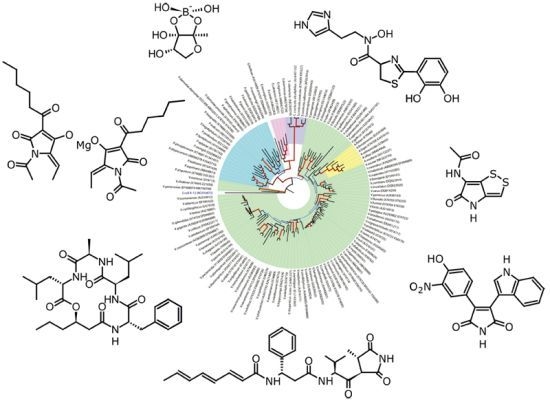
- Evolutionary relationship of Vibrionaceae species. The evolutionary history was inferred using the Neighbor-Joining method [4]. The optimal tree with the sum of branch length = 1.64144427 is shown. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) are shown next to the brances [5]. The tree is drawn to scale, with branch lengths in the same units as those if the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Jukes-Cantor method [6] and are in the units of the number of base substitutions per site. The rate variation among sites was modeled with a gamma distribution (shape parameter = 0.46). The analysis involved 127 nucleotide sequences. All ambiguous positions were removed for each sequence pair. There were a total of 1596 positions in the final dataset. Evolutionary analyses were conducted in MEGA5 [7]. Dendogram was made with the Figtree v. 1.3.1 program ( http://tree.bio.ed.ac.uk/) where line color and width are related to the bootstrap support.
- Saitou, N; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol 1987, 4, 406–425. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar]
- Jukes, TH; Cantor, CR. Mammalian Protein Metabolism; Munro, HN, Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132. [Google Scholar]
- Tamura, K; Peterson, D; Stecher, G; Nei, M; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011. [Google Scholar] [CrossRef]
- Thompson, CC; Vicente, ACP; Souza, RC; Vasconcelos, ATR; Vesth, T; Alves, N; Ussery, DW; Iida, T; Thompson, FL. Genomic taxonomy of vibrios. BMC Evol. Biol 2009, 9, 258. [Google Scholar]
- Grimes, DJ; Johnson, CN; Dillon, KS; Flowers, AR; Noriea, NF; Berutti, T. What genomic sequence information has revealed about vibrio ecology in the ocean—A review. Microb. Ecol 2009, 58, 447–460. [Google Scholar]
- Colwell, R. Vibrios in the Environment; John Wiley & Sons, Inc: Hoboken, NJ, USA, 1984. [Google Scholar]
- Faruque, SM; Albert, MJ; Mekalanos, JJ. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol. Mol. Biol. Rev 1998, 62, 1301–1314. [Google Scholar]
- Linkous, DA; Oliver, JD. Pathogenesis of Vibrio vulnificus. FEMS Microbiol. Lett 1999, 174, 207–214. [Google Scholar]
- Blake, PA; Weaver, RE; Hollis, DG. Diseases of humans (other than cholera) caused by Vibrios. Annu. Rev.Microbiol 1980, 34, 341–367. [Google Scholar]
- Thompson, FL. The Biology of Vibrios; ASM Press: Washington, DC, USA, 2006. [Google Scholar]
- Ruby, EG. Lessons from a cooperative, bacterial-animal association: The Vibrio fischeri Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol 1996, 50, 591–624. [Google Scholar]
- Nyholm, SV; McFall-Ngai, MJ. The Winnowing: Establishing the squid-vibrio symbiosis. Nat. Rev. Microbiol 2004, 2, 632–642. [Google Scholar]
- Milton, DL. Quorum sensing in vibrios: Complexity for diversification. Int. J. Med. Microbiol 2006, 296, 61–71. [Google Scholar]
- Islam, MS; Mahmuda, S; Morshed, MG; Bakht, HBM; Khan, MNH; Sack, RB; Sack, DA. Role of Cyanobacteria in the Persistence of Vibrio cholerae O139 in saline microcosms. Can. J. Microbiol 2004, 50, 127–131. [Google Scholar]
- Pruzzo, C; Vezzulli, L; Colwell, RR. Global impact of Vibrio cholerae interactions with chitin. Environ. Microbiol 2008, 10, 1400–1410. [Google Scholar]
- Su, YC; Liu, CC. Vibrio parahaemolyticus: A concern of seafood safety. Food Microbiol 2007, 24, 549–558. [Google Scholar]
- Faruque, SM; Biswas, K; Udden, SMN; Ahmad, QS; Sack, DA; Nair, GB; Mekalanos, JJ. Transmissibility of cholera: In vivo-formed biofilms and their relationship to infectivity and persistence in the environment. Proc. Natl. Acad. Sci. USA 2006, 103, 6350–6355. [Google Scholar]
- Yildiz, FH; Visick, KL. Vibrio biofilms: So much the same yet so different. Trends Microbiol 2009, 17, 109–118. [Google Scholar]
- Yip, ES; Geszvain, K; DeLoney-Marino, CR; Visick, KL. The symbiosis regulator rscs controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol. Microbiol 2006, 62, 1586–1600. [Google Scholar]
- Nyholm, SV; Stabb, EV; Ruby, EG; McFall-Ngai, MJ. Establishment of an animal-bacterial association: Recruiting symbiotic vibrios from the environment. Proc. Natl. Acad. Sci. USA 2000, 97, 10231–10235. [Google Scholar]
- Bayles, KW. The biological role of death and lysis in biofilm development. Nat. Rev. Microbiol 2007, 5, 721–726. [Google Scholar]
- Heidelberg, JF; Heidelberg, KB; Colwell, RR. Bacteria of the gamma-subclass proteobacteria associated with zooplankton in chesapeake bay. Appl. Environ. Microbiol 2002, 68, 5498–5507. [Google Scholar]
- Hunt, DE; Gevers, D; Vahora, NM; Polz, MF. Conservation of the chitin utilization pathway in the Vibrionaceae. Appl. Environ. Microbiol 2008, 74, 44–51. [Google Scholar]
- Svitil, AL; Chadhain, SMN; Moore, JA; Kirchman, DL. Chitin degradation proteins produced by the marine bacterium Vibrio harveyi growing on different forms of chitin. Appl. Environ. Microbiol 1997, 63, 408–413. [Google Scholar]
- Wietz, M; Mansson, M; Gram, L. Chitin stimulates production of the antibiotic andrimid in a Vibrio coralliilyticus strain. Environ Microbiol Rep 2011. 10.11117j.1758-2229.2011.00259.x.. [Google Scholar]
- Goecke, F; Labes, A; Wiese, J; Imhoff, JF. chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser 2010, 409, 267–299. [Google Scholar]
- Riemann, L; Azam, F. Widespread N-acetyl-d-glucosamine uptake among pelagic marine bacteria and its ecological implications. Appl. Environ. Microbiol 2002, 68, 5554–5562. [Google Scholar]
- Okada, K; Iida, T; Kita-Tsukamoto, K; Honda, T. Vibrios commonly possess two chromosomes. J. Bacteriol 2005, 187, 752–757. [Google Scholar]
- Kirkup, BC; Chang, LA; Chang, S; Gevers, D; Polz, MF. Vibrio chromosomes share common history. BMC Microbiol 2010, 10, 137. [Google Scholar]
- Tagomori, K; Iida, T; Honda, T. comparison of genome structures of vibrios, bacteria possessing two chromosomes. J. Bacteriol 2002, 184, 4351–4358. [Google Scholar]
- Hazen, TH; Pan, L; Gu, JD; Sobecky, PA. The contribution of mobile genetic elements to the evolution and ecology of vibrios. FEMS Microbiol. Ecol 2010, 74, 485–499. [Google Scholar]
- Rowe-Magnus, DA; Guerout, AM; Mazel, D. Super-integrons. Res. Microbiol 1999, 150, 641–651. [Google Scholar]
- Mazel, D. Integrons: Agents of bacterial evolution. Nat. Rev. Microbiol 2006, 4, 608–620. [Google Scholar]
- Meibom, KL; Blokesch, M; Dolganov, NA; Wu, CY; Schoolnik, GK. Chitin induces natural competence in Vibrio cholerae. Science 2005, 310, 1824–1827. [Google Scholar]
- Gulig, PA; Tucker, MS; Thiaville, PC; Joseph, JL; Brown, RN. User friendly cloning coupled with chitin-based natural transformation enables rapid mutagenesis of Vibrio vulnificus. Appl. Environ. Microbiol 2009, 75, 4936–4949. [Google Scholar]
- Pollack-Berti, A; Wollenberg, MS; Ruby, EG. Natural transformation of Vibrio fischeri requires tfoX and tfoY. Environ. Microbiol 2010, 12, 2302–2311. [Google Scholar]
- Thompson, FL; Gevers, D; Thompson, CC; Dawyndt, P; Naser, S; Hoste, B; Munn, CB; Swings, J. Phylogeny and molecular identification of vibrios on the basis of multilocus sequence analysis. Appl. Environ. Microbiol 2005, 71, 5107–5115. [Google Scholar]
- Pascual, J; Macian, MC; Arahal, DR; Garay, E; Pujalte, MJ. Multilocus sequence analysis of the central clade of the genus vibrio by using the 16S rRNA, recA, pyrH, rpoD, gyrB, rctB and toxR genes. Int. J. Syst. Evol. Microbiol 2010, 60, 154–165. [Google Scholar]
- Dieckmann, R; Strauch, E; Alter, T. Rapid identification and characterization of vibrio species using whole-cell MALDI-TOF mass spectrometry. J. Appl. Microbiol 2010, 109, 199–211. [Google Scholar]
- Hazen, TH; Martinez, RJ; Chen, YF; Lafon, PC; Garrett, NM; Parsons, MB; Bopp, CA; Sullards, MC; Sobecky, PA. Rapid identification of Vibrio parahaemolyticus by whole-cell matrix-assisted laser desorption ionization-time of flight mass spectrometry. Appl. Environ. Microbiol 2009, 75, 6745–6756. [Google Scholar]
- Wietz, M; Mansson, M; Gotfredsen, CH; Larsen, TO; Gram, L. Antibacterial compounds from marine Vibrionaceae isolated on a global expedition. Mar. Drugs 2010, 8, 2946–2960. [Google Scholar]
- Oclarit, JM; Okada, H; Ohta, S; Kaminura, K; Yamaoka, Y; Iizuka, T; Miyashiro, S; Ikegami, S. Anti-bacillus substance in the marine sponge, Hyatella species, produced by an associated Vibrio species bacterium. Microbios 1994, 78, 7–16. [Google Scholar]
- Yao, CBFF; Al Zereini, W; Fotso, S; Anke, H; Laatsch, H. Aqabamycins A–G: Novel nitro maleimides from a marine Vibrio species: II. Structure elucidation. J. Antibiot 2010, 63, 303–308. [Google Scholar]
- Al-Zereini, W; Yao, CBFF; Laatsch, H; Anke, H. Aqabamycins A–G: Novel nitro maleimides from a marine Vibrio species: I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot 2010, 63, 297–301. [Google Scholar]
- Sato, A. Annual Report Sankyo Research Laboratories; Daiichi Sankyo: Tokyo, Japan, 1995. [Google Scholar]
- Gerber, NN. Cycloprodigiosin from Beneckea Gazogenes. Tetrahedron Lett 1983, 24, 2797–2798. [Google Scholar]
- Elyakov, GB; Kuznetsova, T; Mikhailov, VV; Maltsev, II; Voinov, VG; Fedoreyev, SA. Brominated diphenyl ethers from a marine bacterium associated with the sponge Dysidea sp. Experientia 1991, 47, 632–633. [Google Scholar]
- Sionov, E; Roth, D; Sandovsky-Losica, H; Kashman, Y; Rudi, A; Chill, L; Berdicevsky, I; Segal, E. Antifungal effect and possible mode of activity of a compound from the marine sponge Dysidea herbacea. J. Infect 2005, 50, 453–460. [Google Scholar]
- Bell, R; Carmeli, S; Sar, N. Vibrindole A, a metabolite of the marine bacterium, Vibrio parahaemolyticus, isolated from the toxic mucus of the boxfish Ostracion cubicus. J. Nat. Prod 1994, 57, 1587–1590. [Google Scholar]
- Veluri, R; Oka, I; Wagner-Döbler, I; Laatsch, H. New indole alkaloids from the north sea bacterium Vibrio parahaemolyticus Bio249. J. Nat. Prod 2003, 66, 1520–1523. [Google Scholar]
- Imamura, N; Adachi, K; Sano, H. Magnesidin A, a component of marine antibiotic magnesidin, produced by Vibrio gazogenes Atcc29988. J. Antibiot 1994, 47, 257–261. [Google Scholar]
- Laatsch, H. AntiBase 2010; Wiley-VCH: Weinheim, Germany, 2010. [Google Scholar]
- Adachi, K; Kawabata, Y; Kasai, H; Katsuta, M; Shizuri, Y. Novel ngercheumicin or its salt useful for treating infection caused by Pseudovibrio denitrificans. Patent JP2007230911-A, September 2007. [Google Scholar]
- Imamura, N; Nishijima, M; Takadera, T; Adachi, K; Sakai, M; Sano, H. New anticancer antibiotics pelagiomicins, produced by a new marine bacterium Pelagiobacter variabilis. J. Antibiot 1997, 50, 8–12. [Google Scholar]
- Singh, MP; Menendez, AT; Petersen, PJ; Ding, WD; Maiese, WM; Greenstein, M. Biological and mechanistic activities of phenazine antibiotics produced by culture LL-14I352. J. Antibiot 1997, 50, 785–787. [Google Scholar]
- Daoust, JY; Gerber, NN. Isolation and purification of prodigiosin from Vibrio psychroerythrus. J. Bacteriol 1974, 118, 756–757. [Google Scholar]
- Harwood, CS. Beneckea gazogenes sp. nov., a red, facultatively anaerobic, marine bacterium. Curr. Microbiol 1978, 1, 233–238. [Google Scholar]
- Shieh, WY; Chen, YW; Chaw, SM; Chiu, HH. Vibrio ruber sp. nov., a red, facultatively anaerobic, marine bacterium isolated from sea water. Int. J. Syst. Evol. Microbiol 2003, 53, 479–484. [Google Scholar]
- Oku, N; Kawabata, K; Adachi, K; Katsuta, A; Shizuri, Y. Unnarmicins A and C, new antibacterial depsipeptides produced by marine bacterium Photobacterium sp. MBIC06485. J. Antibiot 2008, 61, 11–17. [Google Scholar]
- Jalal, MAF; Hossain, MB; Vanderhelm, D; Sandersloehr, J; Actis, LA; Crosa, JH. Structure of anguibactin, a unique plasmid-related bacterial siderophore from the fish pathogen Vibrio anguillarum. J. Am. Chem. Soc 1989, 111, 292–296. [Google Scholar]
- Sandy, M; Han, A; Blunt, J; Munro, M; Haygood, M; Butler, A. Vanchrobactin and anguibactin siderophores produced by Vibrio sp. DS40M4. J. Nat. Prod 2010, 73, 1038–1043. [Google Scholar]
- Haygood, MG; Holt, PD; Butler, A. Aerobactin production by a planktonic marine Vibrio sp. Limnol. Oceanogr 1993, 38, 1091–1097. [Google Scholar]
- Martinez, JS; Carter-Franklin, JN; Mann, EL; Martin, JD; Haygood, MG; Butler, A. Structure and membrane affinity of a suite of amphiphilic siderophores produced by a marine bacterium. Proc. Natl. Acad. Sci. USA 2003, 100, 3754–3759. [Google Scholar]
- Yamamoto, S; Okujo, N; Fujita, Y; Saito, M; Yoshida, T; Shinoda, S. Structures of two polyamine containing catecholate siderophores from Vibrio fluvialis. J. Biochem 1993, 113, 538–544. [Google Scholar]
- Takahashi, A; Nakamura, H; Kameyama, T; Kurasawa, S; Naganawa, H; Okami, Y; Takeuchi, T; Umezawa, H. Bisucaberin, a new siderophore, sensitizing tumor-cells to macrophage-mediated cytolysis. 2. Physicochemical properties and structure determination. J. Antibiot 1987, 40, 1671–1676. [Google Scholar]
- Winkelmann, G; Schmid, DG; Nicholson, G; Jung, G; Colquhoun, DJ. Bisucaberin—A dihydroxamate siderophore isolated from Vibrio salmonicida, an important pathogen of farmed Atlantic salmon (Salmo salar). Biometals 2002, 15, 153–160. [Google Scholar]
- Soengas, RG; Anta, C; Espada, A; Paz, V; Ares, IR; Balado, M; Rodriguez, J; Lemos, ML; Jimenez, C. Structural characterization of vanchrobactin, a new catechol siderophore produced by the fish pathogen Vibrio anguillarum serotype O2. Tetrahedron Lett 2006, 47, 7113–7116. [Google Scholar]
- Griffiths, GL; Sigel, SP; Payne, SM; Neilands, JB. Vibriobactin, a siderophore from Vibrio cholerae. J. Biol. Chem 1984, 259, 383–385. [Google Scholar]
- Yamamoto, S; Okujo, N; Yoshida, T; Matsuura, S; Shinoda, S. structure and iron transport activity of vibrioferrin, a new siderophore of Vibrio parahaemolyticus. J. Biochem 1994, 115, 868–874. [Google Scholar]
- Okujo, N; Saito, M; Yamamoto, S; Yoshida, T; Miyoshi, S; Shinoda, S. Structure of vulnibactin, a new polyamine-containing siderophore from Vibrio vulnificus. Biometals 1994, 7, 109–116. [Google Scholar]
- Hill, RT; Hamann, MT; Enticknap, JJ; Rao, KV. Kahalalide-Producing Bacteria. May 2005. Available online: http://www.wipo.int/patentscope/search/en/WO2005042720 accessed on 16 August 2011.
- Chen, X; Schauder, S; Potier, N; van Dorsselaer, A; Pelczer, I; Bassler, BL; Hughson, FM. Structural identification of a bacterial quorum-sensing signal containing boron. Nature 2002, 415, 545–549. [Google Scholar]
- Kuo, A; Blough, NV; Dunlap, PV. Multiple N-acyl-l-homoserine lactone autoinducers of luminescence in the marine symbiotic bacterium Vibrio fischeri. J. Bacteriol 1994, 176, 7558–7565. [Google Scholar]
- Cao, JG; Meighen, EA. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J. Biol. Chem 1989, 264, 21670–21676. [Google Scholar]
- Milton, DL; Chalker, VJ; Kirke, D; Hardman, A; Camara, M; Williams, P. The luxM homologue vanM from Vibrio anguillanrum directs the synthesis of N-(3-hydroxyhexanoyl) homoserine Lactone and N-hexanoylhomoserine lactone. J. Bacteriol 2001, 183, 3537–3547. [Google Scholar]
- De Nys, R; Kumar, N; Sharara, KA; Srinivasan, S; Ball, G; Kjelleberg, S. A new metabolite from the marine bacterium Vibrio angustum S14. J. Nat. Prod 2001, 64, 531–532. [Google Scholar]
- Milton, DL; Hardman, A; Camara, M; Chhabra, SR; Bycroft, BW; Stewart, GSAB; Williams, P. Quorum sensing in Vibrio anguillarum: Characterization of the vanI/vanR locus and identification of the autoinducer N-(3-oxodecanoyl)-l-homoserine lactone. J. Bacteriol 1997, 179, 3004–3012. [Google Scholar]
- Eberhard, A; Burlingame, AL; Eberhard, C; Kenyon, GL; Nealson, KH; Oppenheimer, NJ. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 1981, 20, 2444–2449. [Google Scholar]
- Mansson, M; Nielsen, A; Kjærulff, L; Gotfredsen, CH; Wietz, M; Ingmer, H; Gram, L; Larsen, TO. Inhibition of virulence gene expression in Staphylococcus aureus by novel depsipeptides from a marine Photobacterium. Mar Drugs 2011. submitted for publication. [Google Scholar]
- Noguchi, T; Hwang, DF; Arakawa, O; Sugita, H; Deguchi, Y; Shida, Y; Hashimoto, K. Vibrio alginolyticus, a tetrodotoxin-producing bacterium, in the intestines of the fish Fugu-Vermicularis vermicularis. Mar. Biol 1987, 94, 625–630. [Google Scholar]
- Lee, MJ; Jeong, DY; Kim, WS; Kim, HD; Kim, CH; Park, WW; Park, YH; Kim, KS; Kim, HM; Kim, DS. A tetrodotoxin-producing Vibrio strain, LM-1, from the puffer fish Fugu vermicularis radiatus. Appl. Environ. Microbiol 2000, 66, 1698–1701. [Google Scholar]
- Noguchi, T; Ali, AE; Arakawa, O; Miyazawa, K; Kanoh, S; Shida, Y; Nishio, S; Hashimoto, K. Tetrodonic acid-like substance-a possible precursor of tetrodotoxin. Toxicon 1991, 29, 845–855. [Google Scholar]
- Noguchi, T; Jeon, JK; Arakawa, O; Sugita, H; Deguchi, Y; Shida, Y; Hashimoto, K. Occurrence of tetrodotoxin and anhydrotetrodotoxin in Vibrio sp isolated from the intestines of a xanthid crab, Atergatis floridus. J. Biochem 1986, 99, 311–314. [Google Scholar]
- Suzuki, A; Goto, M. Photolumazines, new naturally occurring inhibitors of riboflavin synthetase. Biochim. Biophys. Acta 1973, 313, 229–234. [Google Scholar]
- Gutierrez, CK; Matsui, GY; Lincoln, DE; Lovell, CR. Production of the phytohormone indole-3-acetic acid by estuarine species of the genus Vibrio. Appl. Environ. Microbiol 2009, 75, 2253–2258. [Google Scholar]
- Matsuura, S; Odaka, M; Sugimoto, T; Goto, T. The structure of pteridines from Photobacterium phosphorium. Chem. Lett 1973, 2, 343–346. [Google Scholar]
- Gram, L; Melchiorsen, J; Bruhn, JB. Antibacterial activity of marine culturable bacteria collected from a global sampling of ocean surface waters and surface swabs of marine organisms. Mar. Biotechnol 2010, 12, 439–451. [Google Scholar]
- Long, RA; Azam, F. Antagonistic interactions among marine pelagic bacteria. Appl. Environ. Microbiol 2001, 67, 4975–4983. [Google Scholar]
- Long, RA; Rowley, DC; Zamora, E; Liu, JY; Bartlett, DH; Azam, F. Antagonistic interactions among marine bacteria impede the proliferation of Vibrio cholerae. Appl. Environ. Microbiol 2005, 71, 8531–8536. [Google Scholar]
- Hibbing, ME; Fuqua, C; Parsek, MR; Peterson, SB. Bacterial competition: Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol 2010, 8, 15–25. [Google Scholar]
- Rypien, KL; Ward, JR; Azam, F. Antagonistic interactions among coral-associated bacteria. Environ. Microbiol 2010, 12, 28–39. [Google Scholar]
- Pohlmann, J; Lampe, T; Shimada, M; Nell, PG; Pernerstorfer, J; Svenstrup, N; Brunner, NA; Schiffer, G; Freiberg, C. Pyrrolidinedione derivatives as antibacterial agents with a novel mode of action. Bioorg. Med. Chem. Lett 2005, 15, 1189–1192. [Google Scholar]
- Singh, MP; Mroczenski-Wildey, MJ; Steinberg, DA; Andersen, RJ; Maiese, WM; Greenstein, M. Biological activity and mechanistic studies of andrimid. J. Antibiot 1997, 50, 270–273. [Google Scholar]
- Fredenhagen, A; Tamura, SY; Kenny, PTM; Komura, H; Naya, Y; Nakanishi, K; Nishiyama, K; Sugiura, M; Kita, H. Andrimid, a new peptide antibiotic produced by an intracellular bacterial symbiont isolated from a brown planthopper. J. Am. Chem. Soc 1987, 109, 4409–4411. [Google Scholar]
- Needham, J; Kelly, MT; Ishige, M; Andersen, RJ. Andrimid and moiramides A–C, metabolites produced in culture by a marine isolate of the bacterium Pseudomonas fluorescens: Structure elucidation and biosynthesis. J. Org. Chem 1994, 59, 2058–2063. [Google Scholar]
- Jin, M; Fischbach, MA; Clardy, J. A biosynthetic gene cluster for the acetyl-CoA carboxylase inhibitor andrimid. J. Am. Chem. Soc 2006, 128, 10660–10661. [Google Scholar]
- Fischbach, MA; Walsh, CT; Clardy, J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc. Natl. Acad. Sci. USA 2008, 105, 4601–4608. [Google Scholar]
- Liu, XY; Fortin, PD; Walsh, CT. Andrimid producers encode an acetyl-CoA carboxyltransferase subunit resistant to the action of the antibiotic. Proc. Natl. Acad. Sci. USA 2008, 105, 13321–13326. [Google Scholar]
- Kenig, M; Reading, C. Holomycin and an antibiotic (Mm-19290) related to tunicamycin, metabolites of Streptomyces clavuligerus. J. Antibiot 1979, 32, 549–554. [Google Scholar]
- Ettlinger, L; Gaumann, E; Hutter, R; Kellerschierlein, W; Kradolfer, F; Neipp, L; Prelog, V; Zahner, H. Stoffwechselprodukte von actinomyceten. 17. Holomycin. Helv. Chim. Acta 1959, 42, 563–569. [Google Scholar]
- Hou, YH; Li, FC; Wang, SJ; Qin, S; Wang, QF. Intergeneric conjugation in holomycin-producing marine Streptomyces sp. strain M095. Microbiol. Res 2008, 163, 96–104. [Google Scholar]
- Li, B; Walsh, CT. Identification of the gene cluster for the dithiolopyrrolone antibiotic holomycin in Streptomyces clavuligerus. Proc. Natl. Acad. Sci. USA 2010, 107, 19731–19735. [Google Scholar]
- Oliva, B; O’Neill, A; Wilson, JM; O’Hanlon, PJ; Chopra, I. Antimicrobial properties and mode of action of the pyrrothine holomycin. Antimicrob. Agents Chemother 2001, 45, 532–539. [Google Scholar]
- Winkler, R; Hertweck, C. Biosynthesis of nitro compounds. ChemBioChem 2007, 8, 973–977. [Google Scholar]
- Vynne, NG; Mansson, M; Nielsen, KF; Gram, L. Bioactivity, chemical profiling, and 16S rRNA based phylogeny of Pseudoalteromonas strains collected on a global research cruise. Mar Biotechnol 2011. [Google Scholar] [CrossRef]
- Gerber, NN; Gauthier, MJ. New prodigiosin-like pigment from Alteromonas rubra. Appl. Environ. Microbiol 1979, 37, 1176–1179. [Google Scholar]
- Kim, D; Lee, JS; Park, YK; Kim, JF; Jeong, H; Oh, TK; Kim, BS; Lee, CH. Biosynthesis of antibiotic prodiginines in the marine bacterium Hahella chejuensis KCTC 2396. J. Appl. Microbiol 2007, 102, 937–944. [Google Scholar]
- Bennett, JW; Bentley, R. Seeing red: The story of prodigiosin. Adv. Appl. Microbiol 2000, 47, 1–32. [Google Scholar]
- Tsao, SW; Rudd, BAM; He, XG; Chang, CJ; Floss, HG. Identification of a red pigment from Streptomyces coelicolor A3(2) as a mixture of prodigiosin derivatives. J. Antibiot 1985, 38, 128–131. [Google Scholar]
- Gerber, NN; Lechevalier, MP. Prodiginine (prodigiosin-like) pigments from Streptomyces and other aerobic Actinomycetes. Can. J. Microbiol 1976, 22, 658–667. [Google Scholar]
- Laatsch, H; Thomson, RH. A revised structure for cycloprodigiosin. Tetrahedron Lett 1983, 24, 2701–2704. [Google Scholar]
- Pandey, R; Chander, R; Sainis, KB. Prodigiosins as anti cancer agents: Living upto their name. Curr. Pharm. Des 2009, 15, 732–741. [Google Scholar]
- Perez-Tomas, R; Vinas, M. New insights on the antitumoral properties of prodiginines. Curr. Med. Chem 2010, 17, 2222–2231. [Google Scholar]
- Williamson, NR; Fineran, PC; Gristwood, T; Chawrai, SR; Leeper, FJ; Salmond, GPC. Anticancer and immunosuppressive properties of bacterial prodiginines. Future Microbiol 2007, 2, 605–618. [Google Scholar]
- Furstner, A. Chemistry and biology of roseophilin and the prodigiosin alkaloids: A survey of the last 2500 years. Angew. Chem. Int. Ed 2003, 42, 3582–3603. [Google Scholar]
- Staric, N; Danevcic, T; Stopar, D. Vibrio sp. DSM 14379 pigment production—a competitive advantage in the environment? Microb. Ecol 2010, 60, 592–598. [Google Scholar]
- Gandhi, NM; Nazareth, J; Divekar, PV; Kohl, H; Desouza, NJ. Magnesidin, a novel magnesium-containing antibiotic. J. Antibiot 1973, 26, 797–798. [Google Scholar]
- Bhat, SV; Kohl, H; Ganguli, BN; Desouza, NJ. Magnesidin-related tetramic acids-synthesis and structural requirements for antibacterial activity. Eur. J. Med. Chem 1977, 12, 53–57. [Google Scholar]
- Kohl, H; Bhat, SV; Patell, JR; Gandhi, NM; Nazareth, J; Divekar, PV; Souza, NJD; Berschei, HG; Fehlhabe, HW. Structure of magnesidin, a new magnesium-containing antibiotic from Pseudomonas magnesiorubra. Tetrahedron Lett 1974, 12, 983–986. [Google Scholar]
- Shieh, WY; Lin, YT; Jean, WD. Pseudovibrio denitrificans gen. nov., sp. nov., a marine, facultatively anaerobic, fermentative bacterium capable of denitrification. Int. J. Syst. Evol. Microbiol 2004, 54, 2307–2312. [Google Scholar]
- De Carvalho, CCCR; Fernandes, P. Production of metabolites as bacterial responses to the marine environment. Mar. Drugs 2010, 8, 705–727. [Google Scholar]
- Hider, RC; Kong, XL. Chemistry and biology of siderophores. Nat. Prod. Rep 2010, 27, 637–657. [Google Scholar]
- Actis, LA; Fish, W; Crosa, JH; Kellerman, K; Ellenberger, SR; Hauser, FM; Sandersloehr, J. Characterization of anguibactin, a novel siderophore from Vibrio anguillarum 775(Pjm1). J. Bacteriol 1986, 167, 57–65. [Google Scholar]
- Lemos, ML; Balado, M; Osorio, CR. Anguibactin- versus vanchrobactin-mediated iron uptake in Vibrio anguillarum: Evolution and ecology of a fish pathogen. Environ. Microbiol. Rep 2010, 2, 19–26. [Google Scholar]
- Di Lorenzo, M; Poppelaars, S; Stork, M; Nagasawa, M; Tolmasky, ME; Crosa, JH. A nonribosomal peptide synthetase with a novel domain organization is essential for siderophore biosynthesis in Vibio anguillarum. J. Bacteriol 2004, 186, 7327–7336. [Google Scholar]
- Naka, H; Lopez, CS; Crosa, JH. Reactivation of the vanchrobactin siderophore system of Vibrio anguillarum by removal of a chromosomal insertion sequence originated in plasmid pJM1 encoding the anguibactin siderophore system. Environ. Microbiol 2008, 10, 265–277. [Google Scholar]
- Kadi, N; Song, LJ; Challis, GL. Bisucaberin biosynthesis: An adenylating domain of the BibC multi-enzyme catalyzes cyclodimerization of N-hydroxy-N-succinylcadaverine. Chem Commun 2008, 5119–5121. [Google Scholar]
- Challis, GL. A widely distributed bacterial pathway for siderophore biosynthesis independent of nonribosomal peptide synthetases. ChemBioChem 2005, 6, 601–611. [Google Scholar]
- Kameyama, T; Takahashi, A; Kurasawa, S; Ishizuka, M; Okami, Y; Takeuchi, T; Umezawa, H. Bisucaberin, a new siderophore, sensitizing tumor-cells to macrophage-mediated cytolysis. 1. Taxonomy of the producing organism, isolation and biological properties. J. Antibiot 1987, 40, 1664–1670. [Google Scholar]
- Bergeron, RJ; Xin, MG; Weimar, WR; Smith, RE; Wiegand, J. Significance of asymmetric sites in choosing siderophores as deferration agents. J. Med. Chem 2001, 44, 2469–2478. [Google Scholar]
- Miller, MJ; Malouin, F. Siderophore-Mediated Drug Delivery: The Design, Synthesis, and Study of Siderophore-Antibiotic and Antifungal Conjugates. In The Development of Iron Chelators for Clinical Use; Bergeron, RJ, Brittenham, GM, Eds.; CRC Press: Boca Raton, FL, USA, 1994; pp. 275–306. [Google Scholar]
- Wittmann, S; Schnabelrauch, M; Scherlitz-Hofmann, I; Mollmann, U; Ankel-Fuchs, D; Heinisch, L. New synthetic siderophores and their beta-lactam conjugates based on diamino acids and dipeptides. Bioorg. Med. Chem 2002, 10, 1659–1670. [Google Scholar]
- Bergeron, RJ; Bharti, N; Singh, S; McManis, JS; Wiegand, J; Green, LG. Vibriobactin antibodies: A vaccine strategy. J. Med. Chem 2009, 52, 3801–3813. [Google Scholar]
- Hamann, MT. Technology Evaluation: Kahalalide F, PharmaMar. Curr. Opin. Mol. Ther 2004, 6, 657–665. [Google Scholar]
- PharmaMar. PharmaMar Licenses Analogs of Kahalalide F to Marinomed for Uses Outside of Oncology/Neurology. Available online: http://www.pharmamar.com/pdf/MarinomedMAR_ENG.pdf accessed on 19 August 2011.
- Matsumura, K. Reexamination of tetrodotoxin production by bacteria. Appl. Environ. Microbiol 1995, 61, 3468–3470. [Google Scholar]
- Tenover, FC; Goering, RV. Methicillin-resistant Staphylococcus aureus strain USA300: Origin and epidemiology. J. Antimicrob. Chemother 2009, 64, 441–446. [Google Scholar]
- Campbell, J; Lin, Q; Geske, GD; Blackwell, HE. New and unexpected insights into the modulation of LuxR-type quorum sensing by cyclic dipeptides. ACS Chem. Biol 2009, 4, 1051–1059. [Google Scholar]
- Unson, MD; Faulkner, DJ. Cyanobacterial symbiont biosynthesis of chlorinated metabolites from Dysidea herbacea (Porifera). Experientia 1993, 49, 349–353. [Google Scholar]
- Laatsch, H. Frontier in Marine Biotechnology; Proksch, P, Ed.; Horizon Bioscience: Norfolk, UK, 2006; pp. 225–288. [Google Scholar]
- Mitova, M; Popov, S; De Rosa, S. Cyclic peptides from a Ruegeria strain of bacteria associated with the sponge Suberites domuncula. J. Nat. Prod 2004, 67, 1178–1181. [Google Scholar]
- Rungprorn, W; Siwu, ERO; Lambert, LK; Dechsakulwatana, C; Barden, MC; Kokpol, U; Blanchfield, JT; Kita, M; Garson, MJ. Cyclic tetrapeptides from marine bacteria associated with the seaweed Diginea sp. and the sponge Halisarca ectofibrosa. Tetrahedron 2008, 64, 3147–3152. [Google Scholar]
- Shin, J; Seo, Y; Lee, HS; Rho, JR; Mo, SJ. A new cyclic peptide from a marine-derived bacterium of the genus Nocardiopsis. J. Nat. Prod 2003, 66, 883–884. [Google Scholar]
- Shaaban, M; Maskey, RP; Wagner-Döbler, I; Laatsch, H. Pharacine, a natural p-cyclophane and other indole derivatives from Cytophaga sp. strain AM13.1. J. Nat. Prod 2002, 65, 1660–1663. [Google Scholar]
- Fischbach, MA. Antibiotics from microbes: Converging to kill. Curr. Opin. Microbiol 2009, 12, 520–527. [Google Scholar]
- Firn, RD; Jones, CG. A darwinian view of metabolism: Molecular properties determine fitness. J. Exp. Bot 2009, 60, 719–726. [Google Scholar]
- Entrez Genome Project Database; National Center of Biotechnology. Available online: http://www.ncbi.nlm.nih.gov/sites/genome accessed on 16 August 2011.














| Bioactivities | Name | Compound class | Source | Other activities | Ref. |
|---|---|---|---|---|---|
| Antibacterial | Andrimid (4) | Pyrrolidinedione | V. coralliilyticus | [45,46] | |
| Aqabamycin A (6) | Nitro maleimide | Vibrio sp. | Anticancer | [47,48] | |
| Aqabamycin B (7) | |||||
| Aqabamycin C (8) | |||||
| Aqabamycin D (9) | |||||
| Aqabamycin E (10) | Maleimide oxime | ||||
| Aqabamycin E’ (11) | |||||
| Aqabamycin F (12) | |||||
| Aqabamycin G (13) | Nitro maleimide | ||||
| B-4607-C | Phenazine | Vibrio sp. | [49] | ||
| Cycloprodigiosin (15) | Prodiginine | V. gazogenes | [50] | ||
| 3,5-Dibromo-2-(3′,5′-dibromo-2′-methoxyphenoxy)-phenol | Diphenyl ether | Vibrio sp. | Antifungal | [51,52] | |
| 2,2-Di-(3-indolyl)-3-indolone | Indole | V. parahaemolyticus | [53,54] | ||
| Griseoluteic acid | Phenazine | Vibrio sp. | [49] | ||
| Holomycin (5) | Pyrrothine | P. halotolerans | [45] | ||
| Indazole-3-carbaldehyde | Indazole | Vibrio sp. | Anticancer | [47] | |
| Magnesidin A (16) | Tetramic acid Mg2+ salt | V. gazogenes | Antialgal | [55] | |
| Moiramide B | Pyrrolidinedione | Vibrio sp. | [56] | ||
| Ngercheumicin A (19) | Depsipeptide | Photobacterium sp. | [57] | ||
| Ngercheumicin B (20) | |||||
| Ngercheumicin C (21) | |||||
| Ngercheumicin D (22) | |||||
| Ngercheumicin E (23) | |||||
| Pelagiomicin C | Phenazine | Vibrio sp. | Anticancer | [49,58,59] | |
| Prodigiosin (14) | Prodiginine | V. psychroerythrus V. gazogenes V. ruber | Antiprotozoan antifungal anticancer | [60–62] | |
| Turbomycin | Indole | Vibrio sp. (V. parahaemolyticus) | Antifungal | [54] | |
| Unnarmicin A (17) | Depsipeptide | Photobacterium sp. | Antifungal | [63] | |
| Unnarmicin C (18) | |||||
| Vibrindole A | Indole | V. parahaemolyticus | Antifungal | [53] | |
| Siderophore | Anguibactin (24) | Catechol hydroxamate | V. anguillarum | Anticancer | [64,65] |
| Aerobactin | Hydroxamate | Vibrio sp. | [66] | ||
| Amphibactin B | Hydroxamate (amphiphilic) | Vibrio sp. | [67] | ||
| Amphibactin C | |||||
| Amphibactin D | |||||
| Amphibactin E | |||||
| Amphibactin F | |||||
| Amphibactin G | |||||
| Amphibactin H | |||||
| Amphibactin I | |||||
| Bis-[3-(2,3-dihydroxybenzoylamino)-propyl]-amin | Catechol | V. fluvialis | [68] | ||
| Bisucaberin (29) | Hydroxamate | V. salmonicida | Anticancer | [69,70] | |
| Divanchrobactin | Catechol | Vibrio sp. | [65] | ||
| Fluvibactin (28) | Catechol Hydroxyphenyloxazolone | V. fluvialis | [66] | ||
| Trivanchrobactin | Catechol | Vibrio sp. | [65] | ||
| Vanchrobactin (25) | Catechol | V. anguillarum | [71] | ||
| Vibriobactin (27) | Catechol Hydroxyphenyloxazolone | V. cholerae | [72] | ||
| Vibrioferrin | Carboxylate | V. parahaemolyticus | [73] | ||
| Vulnibactin (26) | Catechol Hydroxyphenyloxazolone | V. vulnificus | [74] | ||
| Vulnibactin 2 | Vulnibactin precursor | ||||
| Vulnibactin 3 | |||||
| Anticancer | Kahalalide F (30) | Depsipeptide | V. mediterranei (V. shilonii) | Antibacterial antimalarial antifungal | [75] |
| Kahalalide H | [76] | ||||
| Kahalalide J | |||||
| Quorum sensing interference | AI-2 (3) | Furanosyl borate diester | Vibrio | QS | [76] |
| N-hexanoyl-l-homoserine lactone | Homoserine lactone | V. anguillarum | QS | [77] | |
| N-(3-hydroxybutanoyl)-l-homoserine lactone | Homoserine lactone | V. harveyi | QS | [78] | |
| N-(3-hydroxyhexanoyl)-l-homoserine lactone | Homoserine lactone | V. anguillarum | QS | [79] | |
| [1-(2′-methylpropoxy)-2-hydroxy-2-methylpropoxy] butane (41) | P. angustum (V. angustum) | QS | [80] | ||
| N-(3-oxodecanoyl)-l-homoserine lactone | Homoserine lactone | V. anguillarum | QS | [81] | |
| N-(3-oxohexanoyl)-l-homoserine lactone (1) | Homoserine lactone | V. fischeri V. cholerae V. harveyi V. anguillarum | QS | [17,82] | |
| N-octanoyl-l-homoserine lactone (2) | Homoserine lactone | V. fischeri | QS | [77] | |
| Solonamide A (32) | Depsipeptide | P. halotolerans | QSI Gram pos | [83] | |
| Solonamide B (33) | |||||
| Na channel blocker | Anhydro-tetrodotoxin | Vibrio sp. | [84,85] | ||
| 4-epi-tetrodotoxin | Vibrio sp. | [84,85] | |||
| Tetrodonic acid | Vibrio sp. | [85,86] | |||
| Tetrodotoxin (31) | V. harveyi V. alginolyticus V. fischeri | [84,85,87] | |||
| Riboflavin synthase inhibitor | 7-hydroxy-6-methyl-8-(1-d-ribityl)lumazine | Pteridine | P. phosphoreum | [88] | |
| Photolumazine A | |||||
| Photolumazine B | |||||
| Photolumazine C | |||||
| Misc. | Arundine | Indole | V. parahaemolyticus | [54] | |
| Benzoic acid | Aromatic | Vibrio sp. | [47] | ||
| 3,3-Bis-(3-indolyl)butan-2-one | Indole | V. parahaemolyticus | [54] | ||
| 3,3′-Bisindolylmethane | |||||
| 1,4-dithiane | Vibrio sp. | [47] | |||
| 3-hydroxybenzoic acid | Aromatic | Vibrio sp. | [47] | ||
| 4-hydroxycinnamic acid | |||||
| p-Hydroxyphenyl-acetamide | Aromatic | V. parahaemolyticus | [54] | ||
| Indole-3-carboxaldehyde | Indole | V. parahaemolyticus | [53] | ||
| Indole-3-acetic acid | Indole | Vibrio sp. | [89] | ||
| 6-methyl-8-d-ribityl-2,4,7-trioxopteridine | Pteridine | P. phosphoreum | [90] | ||
| 3-nitro-4-hydroxy-benzaldehyde | Nitro aromatic | Vibrio sp. | [47] | ||
| 3-nitro-4-hydroxycinnamic acid | |||||
| 3-nitro-1H-indazole | |||||
| Pharacine (43) | Terephthalic ester | V. parahaemolyticus | [54] | ||
| Phenylacetic acid | Aromatic | Vibrio sp. | [47] | ||
| Phenyl-2-bis-indolylmethane | Indol | ||||
| Photopterin A | Pteridine | P. phosphoreum | [90] | ||
| 8-d-ribityl-2,4,7-trioxopteridine | |||||
| Trisindoline | Indole | V. parahaemolyticus | [54] | ||
| 1,1,3-Tris-(3-indolyl)butane | |||||
| 1,1′,1″-Trisindolyl-methane (42) |
© 2011 by the authors; licensee MDPI, Basel, Switzerland This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Mansson, M.; Gram, L.; Larsen, T.O. Production of Bioactive Secondary Metabolites by Marine Vibrionaceae. Mar. Drugs 2011, 9, 1440-1468. https://doi.org/10.3390/md9091440
Mansson M, Gram L, Larsen TO. Production of Bioactive Secondary Metabolites by Marine Vibrionaceae. Marine Drugs. 2011; 9(9):1440-1468. https://doi.org/10.3390/md9091440
Chicago/Turabian StyleMansson, Maria, Lone Gram, and Thomas O. Larsen. 2011. "Production of Bioactive Secondary Metabolites by Marine Vibrionaceae" Marine Drugs 9, no. 9: 1440-1468. https://doi.org/10.3390/md9091440