Transfer of Multidrug-Resistant Bacteria Between Intermingled Ecological Niches: The Interface Between Humans, Animals and the Environment
Abstract
:1. Introduction
2. Antimicrobial Use in Animals
2.1. Companion Animals
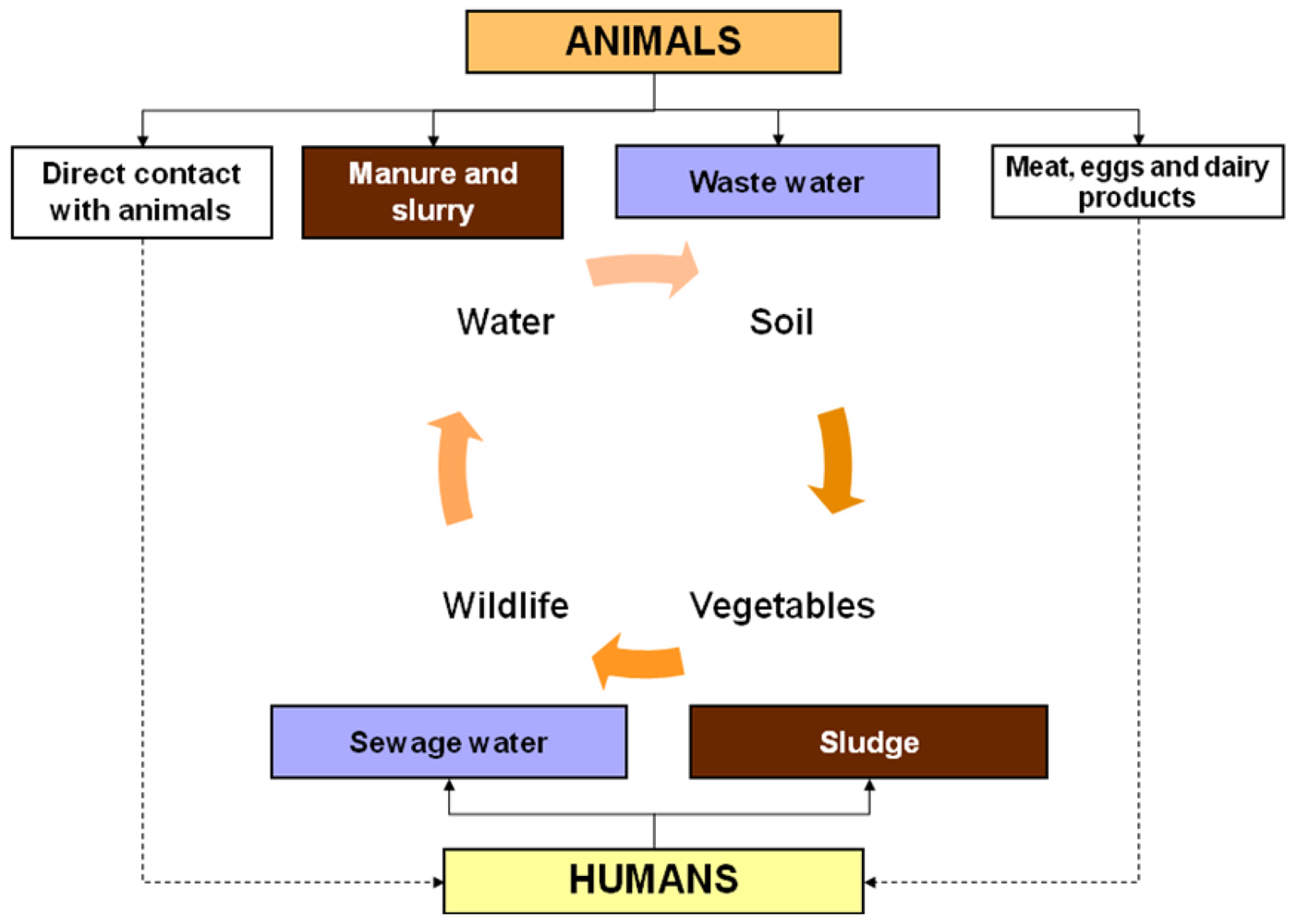
2.2. Production Animals
3. Environmental Dispersion of Antimicrobial Resistance
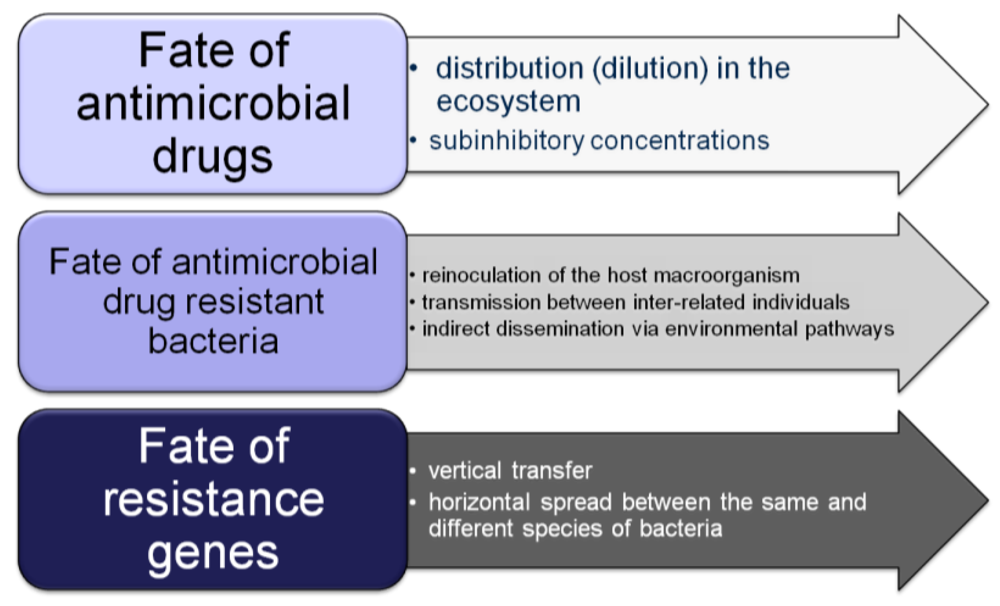
4. Microbial Resistances in Wild Animals
5. Conclusions
Acknowledgements
References
- Schwarz, S.; Kehrenberg, C.; Walsh, T. Use of antimicrobial agents in veterinary medicine and food animal production. Int. J. Antimicrob. Agents 2001, 17, 431–437. [Google Scholar] [CrossRef]
- Rantala, M.; Hölsö, K.; Lillas, A.; Huovinen, P.; Kaartinen, L. Survey of condition-based prescribing of antimicrobial drugs for dogs at a veterinary teaching hospital. Vet. Rec. 2004, 155, 259–262. [Google Scholar] [CrossRef]
- Cabello, F. Heavy use of prophylactic antibiotics in aquaculture: A growing problem for human and animal health and for the environment. Environ. Microbiol. 2006, 8, 1137–1144. [Google Scholar] [CrossRef]
- Sande-Bruinsma, N.; Grundmann, H.; Verloo, D.; Tiemersma, E.; Monen, J.; Goossens, H.; Ferech, M. Antimicrobial drug use and resistance in Europe. Emerg. Infect. Dis. 2008, 11, 1722–1730. [Google Scholar]
- Turnidge, J.; Christiansen, K. Antibiotic use and resistance—Proving the obvious. Lancet 2005, 365, 548–549. [Google Scholar]
- Enne, V. Reducing antimicrobial resistance in the community by restricting prescribing: Can it be done? J. Antimicrob. Chemother. 2010, 65, 179–182. [Google Scholar] [CrossRef]
- Acar, J.; Röstel, B. Antimicrobial resistance: An overview. Rev. Sci. Tech. Off. Int. Epiz. 2001, 20, 797–810. [Google Scholar]
- Memish, Z.; Venkatesh, S.; Shibl, A. Impact of travel on international spread of antimicrobial resistance. Int. J. Antimicrob. Ag. 2003, 21, 135–142. [Google Scholar] [CrossRef]
- Silbergeld, E.; Graham, J.; Price, L. Industrial food animal production, antimicrobial resistance, and human health. Annu. Rev. Public Health 2008, 29, 151–169. [Google Scholar] [CrossRef]
- Barbosa, T.; Levy, S. The impact of antibiotic use on resistance development and persistence. Drug Resist. Update 2000, 3, 303–311. [Google Scholar] [CrossRef]
- Livermore, D. Bacterial resistance: Origins, epidemiology, and impact. Clin. Infect. Dis. 2003, 36, S11–S23. [Google Scholar] [CrossRef]
- McDermott, P.; Walker, R.; White, D. Antimicrobials: Modes of action and mechanisms of resistance. Int. J. Toxicol. 2003, 22, 135–143. [Google Scholar] [CrossRef]
- Smillie, C.; Smith, M.; Friedman, J.; Cordero, O.; David, L.; Alm, E. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011, 480, 241–244. [Google Scholar] [CrossRef]
- Summers, A.O. Generally overlooked fundamentals of bacterial genetics and ecology. Clin. Infect. Dis. 2002, 34 (Suppl. 3), S85–S92. [Google Scholar] [CrossRef]
- O’Brien, T.F. Emergence, spread, and environmental effect of antimicrobial resistance: How use of an antimicrobial anywhere can increase resistance to any antimicrobial anywhere else. Clin. Infect. Dis. 2002, 34 (Suppl. 3), S78–S84. [Google Scholar] [CrossRef]
- Aiello, A.; Larson, E. Antibacterial clearing and hygiene products as an emerging risk factor for antibiotic resistance in the community. Lancet Infect. Dis. 2003, 3, 501–506. [Google Scholar] [CrossRef]
- Hegstad, K.; Langsrud, S.; Lunestad, B.; Scheie, A.; Sunde, M.; Yazdankhah, S. Does the wide use of quaternary ammonium compounds enhance the selection and spread of antimicrobial resistance and thus threaten our health? Microb. Drug Resist. 2010, 16, 91–104. [Google Scholar] [CrossRef]
- McEwen, S.; Fedorka-Cray, P. Antimicrobial use and resistance in animals. Clin. Infect. Dis. 2002, 34 (Suppl. 3), S93–S106. [Google Scholar] [CrossRef]
- Overcoming Antimicrobial Resistance. Report on Infectious Diseases.; World Health Organization: Geneva, Switzerland, 2000; No. WHO/CDS/2000.2.
- Nelson, R. Antibiotic development pipeline runs dry. Lancet 2003, 362, 1726–1727. [Google Scholar] [CrossRef]
- Norrby, S.; Nord, C.; Finch, R. Lack of development of new antimicrobial drugs: A potential serious threat to public health. Lancet Infect. Dis. 2005, 5, 115–119. [Google Scholar] [CrossRef]
- Travers, K.; Barza, M. Morbidity of infections caused by antimicrobial-resistant bacteria. Clin. Infect. Dis. 2002, 34 (Suppl. 3), 131–134. [Google Scholar] [CrossRef]
- Rice, L.B. Emerging issues in the management of infections caused by multidrug-resistant gram-negative bacteria. Cleve Clin. J. Med. 2007, 74 (Suppl. 4), S12–S20. [Google Scholar] [CrossRef]
- Wilcox, M. The tide of antimicrobial resistance and selection. Int. J. Antimicrob. Agents 2009, 34, S6–S10. [Google Scholar] [CrossRef]
- Bohnhoff, M.; Miller, C. 1962 Enhanced susceptibility to Salmonella infection in streptomycin-treated mice. J. Infect. Dis. 2004, 111, 117–127. [Google Scholar] [CrossRef]
- Barza, M.; Travers, K. Excess infections due to antimicrobial resistance: The “Attributable Fraction”. Clin. Infect. Dis. 2002, 34 (Suppl. 3), S126–S130. [Google Scholar] [CrossRef]
- Schwartz, T.; Wolfgang, K.; Jansen, B.; Obst, U. Detection of antibiotic-resistant bacteria and their resistance genes in wastewater, surface water, and drinking water biofilms. FEMS Microbiol. Ecol. 2003, 43, 325–335. [Google Scholar] [CrossRef]
- French, G. Clinical impact and relevance of antibiotic resistance. Adv. Drug Deliv. Rev. 2005, 57, 1514–1527. [Google Scholar] [CrossRef]
- Singer, R.; Finch, R.; Wegener, H.; Bywater, R.; Walters, J.; Lipsitch, M. Antibiotic resistance—The interplay between antibiotic use in animals and human beings. Lancet Infect. Dis. 2003, 3, 47–51. [Google Scholar] [CrossRef]
- Turnidge, J. Antibiotic use in animals-prejudices, perceptions and realities. J. Antimicrob. Chemoth. 2004, 53, 26–27. [Google Scholar] [CrossRef]
- Hammerum, A.; Heuer, O. Human health hazards from antimicrobial-resistant Escherichia coli of animal origin. Clin. Infect. Dis. 2009, 48, 916–921. [Google Scholar] [CrossRef]
- Anderson, A.; Nelson, J.; Rossiter, S.; Angulo, F. Public health consequences of use of antimicrobial agents in food animals in the United States. Microb. Drug Resist. 2003, 9, 373–379. [Google Scholar] [CrossRef]
- Angulo, F.; Nargund, V.; Chiller, T. An evidence of an association between use of anti-microbial agents in food animals and anti-microbial resistance among bacteria isolated from humans and the human health consequences of such resistance. J. Vet. Med. B 2004, 51, 374–379. [Google Scholar] [CrossRef]
- Smith, H.W. Transfer of antibiotic resistance from animal and human strains of Escherichia coli to resistant E. coli in the alimentary tract of man. Lancet 1969, 1, 1174–1176. [Google Scholar]
- Guardabassi, L.; Schwarz, S.; Lloyd, D. Pet animals as reservoirs of antimicrobial-resistant bacteria. J. Antimicrob. Chemoth. 2004, 54, 321–332. [Google Scholar] [CrossRef]
- Malik, S.; Coombs, G.; O’Brien, F.; Peng, H.; Barton, M. Molecular typing of methicillin-resistant staphylococci isolated from cats and dogs. J. Antimicrob. Chemoth. 2006, 58, 428–431. [Google Scholar] [CrossRef]
- Damborg, P.; Top, J.; Hendrickx, A.; Dawson, S.; Willems, R.; Guardabassi, L. Dogs are a reservoir of ampicillin-resistant Enterococcus faecium lineages associated with human infections. Appl. Environ. Microbiol. 2009, 75, 2360–2365. [Google Scholar] [CrossRef]
- Paul, N.; Moodley, A.; Ghibaudo, G.; Guardabassi, L. Carriage of methicillin-resistant Staphylococcus pseudintermedius in small animal veterinarians: indirect evidence of zoonotic transmission. Zoonoses Public Health 2011, 58, 533–539. [Google Scholar] [CrossRef]
- Harada, K.; Okada, E.; Shimizu, T.; Kataoka, Y.; Sawada, T.; Takahashi, T. Antimicrobial resistance, virulence profiles, and phylogenetic groups of fecal Escherichia coli isolates: A comparative analysis between dogs and their owners in Japan. Comp. Immunol. Microbiol. Infect. Dis. 2012, 35, 139–144. [Google Scholar] [CrossRef]
- Kwon, K.; Moon, B.; Hwang, S.; Park, Y. Detection of CC17 Enterococcus faecium in dogs: A comparison with human isolates. Zoonoses Public Health 2012, 59, 375–378. [Google Scholar] [CrossRef]
- Leonard, E.; Pearl, D.; Finley, R.; Janecko, N.; Reid-Smith, R.; Peregrine, A.; Weese, J. Comparison of antimicrobial resistance patterns of Salmonella spp. and Escherichia coli recovered from pet dogs from volunteer households in Ontario (2005–2006). J. Antimicrob. Chemoth. 2012, 67, 174–181. [Google Scholar] [CrossRef]
- Wan, M.; Fu, S.; Lo, Y.; Huang, T.; Cheng, M.; Chou, C. Heterogeneity and phylogenetic relationships of community-associated methicillin-sensitive/resistant Staphylococcus aureus isolates in healthy dogs, cats and their owners. J. Appl. Microbiol. 2012, 112, 205–213. [Google Scholar] [CrossRef]
- Thornton, P.K. Livestock production: Recent trends, future prospects. Phil. Trans. Roy. Soc. B 2010, 365, 2853–2867. [Google Scholar] [CrossRef]
- Tollefson, L.; Karp, B. Human health impact from antimicrobial use in food animals. Med. Infect. Dis. 2004, 34, 514–521. [Google Scholar]
- Love, D.; Davis, M.; Bassett, A.; Gunther, A.; Nachman, K. Dose imprecision and resistance: Free-choice medicated feeds in industrial food animal production in the United States. Environ. Health Perspect. 2011, 119, 279–283. [Google Scholar]
- Dibner, J.; Richards, J. Antibiotic growth promoters in agriculture: History and mode of action. Poultry Sci. 2005, 84, 634–643. [Google Scholar]
- Looft, T.; Johnson, T.A.; Allen, H.K.; Bayles, D.O.; Alt, D.P.; Stedtfeld, R.D.; Sul, W.J.; Stedtfeld, T.M.; Chai, B.; Cole, J.R.; et al. In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. USA 2012, 109, 1691–1696. [Google Scholar]
- Berge, A.; Moore, D.; Sischo, W. F. Field trial evaluating the influence of prophylactic and therapeutic antimicrobial administration on antimicrobial resistance of fecal Escherichia coli in dairy calves. Appl. Environ. Microbiol. 2006, 72, 3872–3878. [Google Scholar] [CrossRef]
- Smith, J.L.; Drum, D.J.V.; Dai, Y.; Kim, J.M.; Sanchez, S.; Maurer, J.J.; Hofacre, C.L.; Lee, M.D. Impact of antimicrobial usage on antimicrobial resistance in commensal Escherichia coli strains colonizing broiler chickens. Appl. Environ. Microbiol. 2007, 73, 1404–1414. [Google Scholar] [CrossRef]
- Da Costa, P.; Oliveira, M.; Bica, A.; Vaz-Pires, P.; Bernardo, F. Effects of antimicrobial treatment on selection of resistant Escherichia coli in broiler fecal flora. Microb. Drug Resist. 2008, 14, 299–306. [Google Scholar] [CrossRef]
- Furtula, V.; Farrell, E.; Diarrassouba, F.; Rempel, H.; Pritchard, J.; Diarra, M. Veterinary pharmaceuticals and antibiotic resistance of Escherichia coli isolates in poultry litter from commercial farms and controlled feeding trials. Poultry Sci. 2010, 89, 180–188. [Google Scholar] [CrossRef]
- Collignon, P.; Powers, J.; Chiller, T.; Aidara-Kane, A.; Aarestrup, F. World Health Organization ranking of antimicrobials according to their importance in human medicine: A critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin. Infect. Dis. 2009, 49, 132–141. [Google Scholar] [CrossRef]
- Fey, P.D.; Safranek, T.J.; Rupp, M.E.; Dunne, E.F.; Ribot, E.; Iwen, P.C.; Bradford, P.A.; Angulo, F.J.; Hinrichs, S.H. Ceftriaxone-resistant Salmonella infection acquired by a child from cattle. New Engl. J. Med. 2000, 342, 1242–1249. [Google Scholar] [CrossRef]
- McDermott, P.; Bodeis, S.M.; English, L.L.; White, D.G.; Walker, R.D.; Zhao, S.; Simjee, S.; Wagner, D.D. Ciprofloxacin resistance in Campylobacter jejuni evolves rapidly in chickens treated with fluoroquinolones. J. Infect. Dis. 2002, 185, 837–840. [Google Scholar] [CrossRef]
- González-Sanz, R.; Herrera-León, S.; de la Fuente, M.; Arroyo, M.; Echeita, M. Emergence of extended-spectrum β-lactamases and AmpC-type β-lactamases in human Salmonella isolated in Spain from 2001 to 2005. J. Antimicrob. Chemoth. 2009, 64, 1181–1186. [Google Scholar] [CrossRef]
- Van den Bogaard, A.; London, N.; Driessen, C.; Stobberingh, E. Antibiotic resistance of faecal Escherichia coli in poultry, poultry farmers and poultry slaughterers. J. Antimicrob. Chemoth. 2001, 47, 763–771. [Google Scholar] [CrossRef]
- Musgrove, M.; Jones, D.; Northcutt, J.; Cox, N.; Harrison, M.; Fedorka-Cray, P.; Ladely, S. Antimicrobial resistance in Salmonella and Escherichia coli isolated from commercial shell eggs. Poultry Sci. 2006, 85, 1665–1669. [Google Scholar]
- Haran, K.; Godden, S.; Boxrud, D.; Jawahir, S.; Bender, J.; Sreevatsan, S. Prevalence and characterization of Staphylococcus aureus, including methicillin-resistant Staphylococcus aureus, isolated from bulk tank milk from Minnesota dairy farm. J. Clin. Microbiol. 2012, 50, 688–695. [Google Scholar]
- Zhao, S.; Blickenstaff, K.; Bodeis-Jones, S.; Gaines, S.; Tong, E.; McDermott, P. Comparison of the prevalences and antimicrobial resistances of Escherichia coli isolates from different retail meats in the United States, 2002 to 2008. Appl. Environ. Microbiol. 2012, 78, 1701–1707. [Google Scholar] [CrossRef]
- Sharma, R.; Larney, F.; Chen, J.; Yanke, L.; Morrison, M.; Topp, E.; McAllister, T.; Yu, Z. Selected antimicrobial resistance during composting of manure from cattle administered sub-therapeutic antimicrobials. J. Environ. Qual. 2009, 38, 567–575. [Google Scholar] [CrossRef]
- Wang, L.; Oda, Y.; Grewal, S.; Morrison, M.; Michel, F., Jr.; Yu, Z. Persistence of resistance to erythromycin and tetracycline in swine manure during simulated composting and lagoon treatments. Microb. Ecol. 2012, 63, 32–40. [Google Scholar] [CrossRef]
- Sapkota, A.; Curriero, F.; Gibson, K.; Schwab, K. Antibiotic-resistant Enterococci and fecal indicators in surface water and groundwater impacted by a concentrated Swine feeding operation. Environ. Health Perspect. 2007, 115, 1040–1045. [Google Scholar] [CrossRef]
- Wegener, H.C. Antibiotics in animal feed and their role in resistance development. Curr. Opin. Microbiol. 2003, 6, 439–445. [Google Scholar] [CrossRef]
- Dancer, S. How antibiotics can make us sick: The less obvious adverse effects of antimicrobial chemotherapy. Lancet Infect. Dis. 2004, 4, 611–619. [Google Scholar] [CrossRef]
- Moulin, G.; Cavalié, P.; Pellanne, I.; Chevance, A.; Laval, A.; Millemann, Y.; Colin, P.; Chauvin, C. A comparison of antimicrobial usage in human and veterinary medicine in France from 1999 to 2005. J. Antimicrob. Chemoth. 2008, 62, 617–625. [Google Scholar] [CrossRef]
- Wright, G. Antibiotic resistance in the environment: A link to the clinic? Curr. Opin. Microbiol. 2010, 13, 589–594. [Google Scholar] [CrossRef]
- Aarestrup, F.; Seyfarth, A.; Emborg, H.; Pedersen, K.; Hendriksen, R.; Bager, F. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal Enterococci from food animals in Denmark. Antimicrob. Agents Chemother. 2001, 45, 2054–2059. [Google Scholar] [CrossRef]
- Impacts of Antimicrobial Growth Promoter Termination in Denmark. The WHO International Review Panel’s Evaluation of the Termination of the Use of Antimicrobial Growth Promoters in Denmark.; World Health Organization: Geneva, Switzerland, 2000; No.: WHO/CDS/CPE/ZFK/ 2003.1.
- Diarra, M.S.; Silversides, F.G.; Diarrassouba, F.; Pritchard, J.; Mason, L.; Brousseau, R.; Bonnet, C.; Delaquis, P.; Bach, S.; Skura, B.J.; Topp, E. Impact of feed supplementation with antimicrobial agents on growth performance of broiler chickens, Clostridium perfringens and Enterococcus counts, and antibiotic resistance phenotypes and distribution of antimicrobial resistance determinants in Escherichia coli isolate. Appl. Environ. Microbiol. 2007, 73, 6566–6576. [Google Scholar] [CrossRef]
- Martins da Costa, P.; Belo, A.; Gonçalves, J.; Bernardo, F. Field trial evaluating changes in prevalence and patterns of antimicrobial resistance among Escherichia coli and Enterococcus spp. isolated from growing broilers medicated with enrofloxacin, apramycin and amoxicillin. Vet. Microbiol. 2009, 139, 284–292. [Google Scholar] [CrossRef]
- Martins da Costa, P.; Bica, A.; Vaz-Pires, P.; Bernardo, F. Effects of antimicrobial treatment on selection of resistant Enterococci in broiler fecal flora. Prev. Vet. Med. 2009, 139, 284–292. [Google Scholar]
- Martinez, J. Natural antibiotic resistance and contamination by antibiotic resistance determinants: The two ages in the evolution of resistance to antimicrobials. Front. Microbiol. 2012, 3, 1–3. [Google Scholar] [CrossRef]
- Iverson, A.; Kühn, I.; Franklin, A.; Möllby, R. High prevalence of vancomycin resistant Enterococci in swedish sewage. Appl. Environ. Microbiol. 2002, 68, 2838–2842. [Google Scholar]
- Reinthaler, F.F.; Posch, J.; Feierl, G.; Wust, G.; Haas, D.; Ruckenbauer, G.; Mascher, F.; Marth, E. Antibiotic resistance of E. coli in sewage and sludge. Water Res. 2003, 37, 1685–1690. [Google Scholar] [CrossRef]
- Da Costa, P.; Vaz-Pires, V.; Bernardo, F. Antimicrobial resistance in Escherichia coli isolated in wastewater and sludge from poultry slaughterhouses wastewater plants. J. Environ. Health 2008, 70, 40–45. [Google Scholar]
- Chagas, T.; Seki, L.; Cury, J.; Oliveira, J.; Dávila, A.; Silva, D.; Asensi, M. Multiresistance, β-lactamase-encoding genes and bacterial diversity in hospital wastewater in Rio de Janeiro, Brazil. J. Appl. Microbiol. 2011, 111, 572–581. [Google Scholar] [CrossRef]
- Lindberg, R.; Jarnheimer, P.; Olsen, B.; Johansson, M.; Tysklind, M. Determination of antibiotic substances in hospital sewage water using solid phase extraction and liquid chromatography/ mass spectrometry and group analogue internal standards. Chemosphere 2004, 57, 1479–1488. [Google Scholar] [CrossRef]
- Lindberg, R.; Wennberg, P.; Johansson, M.; Tysklind, M.; Andersson, B. Screening of human antibiotic substances and determination of weekly mass flows in five sewage treatment plants in Sweden. Environ. Sci. Technol. 2005, 39, 3421–3429. [Google Scholar]
- Watanabe, N.; Bergamaschi, B.; Loftin, K.; Meyer, M.; Harter, T. Use and environmental occurrence of antibiotics in freestall dairy farms with manured forage fields. Environ. Sci. Technol. 2010, 44, 6591–6600. [Google Scholar]
- Haggard, B.; Bartsch, L. Net changes in antibiotic concentrations downstream from an effluent discharge. J. Environ. Qual. 2009, 38, 343–352. [Google Scholar] [CrossRef]
- Massey, L.; Haggard, B.; Galloway, J.; Loftin, K.; Meyer, M.; Green, W. Antibiotic fate and transport in three effluent-dominated Ozark streams. Ecol. Eng. 2010, 36, 930–938. [Google Scholar]
- Tello, A.; Austin, B.; Telfer, T. Selective pressure of antibiotic pollution on bacteria of importance to public health. Environ. Health Perspect. 2012, 120, 1100–1106. [Google Scholar] [CrossRef]
- Aminov, R. The role of antibiotics and antibiotic resistance in nature. Environ. Microbiol. 2009, 1112, 2970–2988. [Google Scholar] [CrossRef]
- Aminov, R. Horizontal gene exchange in environmental microbiota. Front. Microbiol. 2011, 2, 1–19. [Google Scholar] [CrossRef]
- Papandreou, S.; Pagonopoulou, O.; Vantarakis, A.; Papapetropoulou, M. Multiantibiotic resistanceof gram-negative bacteria isolated from drinking water samples in southwest Greece. J. Chemother. 2000, 12, 267–273. [Google Scholar]
- Ash, R.; Mauck, B.; Morgan, M. Antibiotic resistance of Gram-negative bacteria in rivers, United States. Emerg. Infect. Dis. 2002, 8, 713–716. [Google Scholar] [CrossRef]
- Alm, E.; Burke, J.; Spain, A. Fecal indicator bacteria are abundant in wet sand at freshwater beaches. Water Res. 2003, 37, 3978–3982. [Google Scholar] [CrossRef]
- Roe, M.; Veja, E.; Pillai, S. Antimicrobial resistance markers of class 1 and class 2 integron-bearing Escherichia coli from irrigation water and sediments. Emerg. Infect. Dis. 2003, 9, 822–826. [Google Scholar] [CrossRef]
- Johnston, L.; Jaykus, L. Antimicrobial resistance of Enterococcus species isolated from produce. Appl. Environ. Microbiol. 2004, 70, 3133–3137. [Google Scholar] [CrossRef]
- Jackson, W.; Piper, J. The necessary marriage between ecology and agriculture. Ecology 1989, 70, 1091–1993. [Google Scholar] [CrossRef]
- Mills, J. Biodiversity loss and emerging infectious disease: An example from the rodent-borne hemorrhagic fevers. Biodiversity 2006, 7, 9–17. [Google Scholar] [CrossRef]
- Corrêa, A.; Albarnaz, J.; Moresco, V.; Poli, C.; Teixeira, A.; Simões, C.; Barardi, C. Depuration dynamics of oysters (Crassostrea gigas) artificially contaminated by Salmonella enterica serovar Typhimurium. Mar. Environ. Res. 2007, 63, 479–489. [Google Scholar] [CrossRef]
- Canesi, L.; Pruzzo, C.; Tarsi, R.; Gallo, G. Surface interactions between Escherichia coli and hemocytes of the Mediterranean mussel Mytilus galloprovincialis Lam. leading to efficient bacterial clearance. Appl. Environ. Microbiol. 2001, 67, 464–468. [Google Scholar] [CrossRef]
- Zampini, M.; Canesi, L.; Betti, M.; Ciacci, C.; Tarsi, R.; Gallo, G.; Pruzzo, C. Role for mannose-sensitive hemagglutitin in promoting interactions between Vibrio cholerae El Tor and mussel hemolymph. Appl. Environ. Microbiol. 2003, 69, 5711–5715. [Google Scholar] [CrossRef]
- Antunes, F.; Hinzmann, M.; Lopes-Lima, M.; Machado, J.; Martins da Costa, P. Relationship between environmental microbiota and indigenous bacteria found in hemolymph, extrapallial fluid and mucus of Anodonta cygnea (Linnaeus, 1758). Microb. Ecol. 2010, 60, 304–309. [Google Scholar] [CrossRef]
- Cavallo, R.; Acquaviva, M.; Stabili, L. Culturable heterotrophic bacteria in seawater and Mytilus galloprovincialis from a Mediterranean area (Northern Ionian Sea-Italy). Environ. Monit. Assess. 2009, 149, 465–475. [Google Scholar] [CrossRef]
- Middleton, J.; Ambrose, A. Enumeration and antibiotic resistance patterns of fecal indicator organisms isolated from migratory Canada geese (Branta canadensis). J. Wildl. Dis. 2005, 41, 334–341. [Google Scholar]
- Costa, D.; Poeta, P.; Saenz, Y.; Vinue, L.; Rojo-Bezares, B.; Jouini, A.; Zarazaga, M.; Rodrigues, J.; Torres, C. Detection of Escherichia coli harbouring extended-spectrum β-lactamases of the CTX-M, TEM and SHV classes in faecal samples of wild animals in Portugal. J. Antimicrob. Chemoth. 2007, 59, 1311–1312. [Google Scholar] [CrossRef]
- Sjolund, M.; Bonnedahl, J.; Hernandez, J.; Bengtsson, S.; Cederbrant, G.; Pinhassi, J.; Kahlmeter, G.; Olsen, B. Dissemination of multidrug-resistant bacteria into the arctic. Emerg. Infect. Dis. 2008, 14, 70–71. [Google Scholar] [CrossRef]
- Poeta, P.; Radhouani, H.; Pinto, L.; Martinho, A.; Rego, V.; Rodrigues, R.; Gonçalves, A.; Rodrigues, J.; Estepa, V.; Torres, C.; Igrejas, G. Wild boars as reservoirs of extended-spectrum β-lactamase (ESBL) producing Escherichia coli of different phylogenetic groups. J. Basic Microbiol. 2009, 49, 584–588. [Google Scholar] [CrossRef]
- Simões, R.; Ferreira, C.; Gonçalves, J.; Álvares, F.; Rio-Maior, H.; Roque, S.; Brandão, R.; Martins da Costa, P. Occurrence of virulence genes in multidrug-resistant Escherichia coli isolates from Iberian wolves (Canis lupus signatus) in Portugal. Eur. J. Wildl. Res. 2012, 58, 677–684. [Google Scholar] [CrossRef]
- Guenther, S.; Grobbel, M.; Lübke-Becker, A.; Goedecke, A.; Friedrich, N.; Wieler, L.; Ewers, C. Antimicrobial resistance profiles of Escherichia coli from common European wild bird species. Vet. Microbiol. 2010, 29, 219–225. [Google Scholar] [CrossRef]
- Pinto, L.; Radhouani, H.; Coelho, C.; Martins da Costa, P.; Simões, R.; Brandão, R.M.L.; Torres, C.; Igrejas, G.; Poeta, P. Genetic detection of extended-spectrum β-lactamases Escherichia coli in birds of prey from Serra da Estrela Natural Reserve of Portugal. Appl. Environ. Microbiol. 2010, 76, 4118–4120. [Google Scholar]
- Smillie, C.; Smith, M.; Friedman, J.; Cordero, O.; David, L.; Alm, E. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011, 480, 241–244. [Google Scholar] [CrossRef]
- Bonnedahl, J.; Drobni, M.; Gauthier-Clerc, M.; Hernandez, J.; Granholm, S.; Kayser, Y.; Melhus, A.; Kahlmeter, G.; Waldenström, J.; Johansson, A.; Olsen, B. Dissemination of Escherichia coli with CTX-M type ESBL between humans and yellow-legged gulls in the south of France. PLoS ONE 2009. [Google Scholar] [CrossRef]
- Literak, I.; Dolejska, M.; Janoszowska, D.; Hrusakova, J.; Meissner, W.; Rzyska, H.; Bzoma, S.; Cizek, A. Antibiotic-resistant Escherichia coli bacteria, including strains with genes encoding the extended-spectrum β-lactamase and QnrS, in waterbirds on the Baltic Sea Coast of Poland. Appl. Environ. Microbiol. 2010, 76, 8126–8134. [Google Scholar] [CrossRef]
- Simões, R.; Poirel, L.; Martins da Costa, P.; Nordmann, P. Seagulls and beaches as a reservoir for emerging extended spectrum beta-lactamase producers in Escherichia coli. Emerg. Infect. Dis. 2010, 16, 110–112. [Google Scholar]
- Mendonça, N.; Leitão, J.; Manageiro, V.; Ferreira, E.; Caniça, M. Spread of extended-spectrum β-lactamase CTX-M-producing Escherichia coli clinical isolates in community and nosocomial environments in Portugal. Antimicrob. Agents Chemother. 2007, 51, 1946–1955. [Google Scholar] [CrossRef]
- Machado, E.; Coque, T.; Cantón, R.; Baquero, F.; Sousa, J.; Peixe, L. Dissemination of Enterobacteriaceae harboring blaCTX-M-15, blaOXA-1, blaTEM-1 and aac(6')-Ib-cr gene in Portugal. Antimicrob. Agents Chemother. 2006, 50, 3220–3221. [Google Scholar] [CrossRef]
- Biyela, P.; Lin, J.; Bezuidenhout, C. The role of aquatic ecosystems as reservoirs of antibiotic resistant bacteria and antibiotic resistance genes. Water Sci. Technol. 2004, 50, 45–50. [Google Scholar]
- Roque, S.; Alvares, F.; Petrucci-Fonseca, F. Utilization espacio-temporal y habitos alimentarios de un grupo reproductor de lobos en el noroeste de Portugal. Galemys 2001, 23, 179–198. [Google Scholar]
- Maynard, C.; Bekal, S.; Sanschagrin, F.; Levesque, R.; Brousseau, R.; Masson, L.; Larivière, S.; Harel, J. Heterogeneity among virulence and antimicrobial resistance gene profiles of extraintestinal Escherichia coli isolates of animal and human origin. J. Clin. Microbiol. 2004, 42, 5444–5452. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Da Costa, P.M.; Loureiro, L.; Matos, A.J.F. Transfer of Multidrug-Resistant Bacteria Between Intermingled Ecological Niches: The Interface Between Humans, Animals and the Environment. Int. J. Environ. Res. Public Health 2013, 10, 278-294. https://doi.org/10.3390/ijerph10010278
Da Costa PM, Loureiro L, Matos AJF. Transfer of Multidrug-Resistant Bacteria Between Intermingled Ecological Niches: The Interface Between Humans, Animals and the Environment. International Journal of Environmental Research and Public Health. 2013; 10(1):278-294. https://doi.org/10.3390/ijerph10010278
Chicago/Turabian StyleDa Costa, Paulo Martins, Luís Loureiro, and Augusto J. F. Matos. 2013. "Transfer of Multidrug-Resistant Bacteria Between Intermingled Ecological Niches: The Interface Between Humans, Animals and the Environment" International Journal of Environmental Research and Public Health 10, no. 1: 278-294. https://doi.org/10.3390/ijerph10010278




