Cryptosporidium and Giardia in Surface Water: A Case Study from Michigan, USA to Inform Management of Rural Water Systems
Abstract
:1. Introduction
2. Experimental Section
2.1. Case Study Background: Michigan, USA
| Characteristic | Rural | Urban |
|---|---|---|
| Source Water | Groundwater | Surface water |
| Common Water System Type | Private wells | Community systems |
| Dominant Waste Streams | Livestock | Human |
| Drinking Water Regulations | Minimal regulation once wells are constructed | Safe Drinking Water Act requirements enforced |

2.2. Prevalence in the Environment

2.2.1. Field Sampling Sites

2.2.2. Sampling Methods
2.2.3. Genotyping
2.2.4. Statistical Analyses
2.3. Relationship between Disease and Land Use
3. Results and Discussion
3.1. Prevalence in the Rural and Urban Water Environments
3.1.1. Occurrence of Cryptosporidium and Giardia in Rural versus Urban Areas

| Site Location | Sample Collection Dates | Cryptosporidium Concentration (Oocyst/100 L) | Giardia Concentration (Oocyst/100 L) | Cryptosporidium Infectivity (MPN/mL) (95% Confidence Interval) |
|---|---|---|---|---|
| Rice Lake Drain | 10/22/2004 | 2600 | <29 | ND |
| 11/4/2004 | 582.2 | <34.2 | ND | |
| Agricultural Drain Tile into Rice Lake Drain | 11/9/2004 | 30.3 | <15.2 | ND |
| 12/6/2004 | 59,900 | 76.3 | 2.301 [0.1789 : 6.238] | |
| 12/7/2004 | 1250 | 50 | ND | |
| Black Creek at Crockett Rd. | 11/9/2004 | <10.6 | 10.6 | -- |
| 11/17/2004 | <9.7 | <9.7 | -- | |
| 12/6/2004 | <9.26 | <9.26 | -- | |
| 12/7/2004 | 38 | 38 | ND | |
| 12/14/2004 | <28.45 | <28.45 | -- | |
| Stoney Creek 1 (at Senecca Rd.) | 11/17/2004 | 64.6 | 21.5 | ND |
| 12/7/2004 | 1350 | <150 | ND | |
| Stoney Creek 2 (at Gorman Rd.) | 12/7/2004 | 450 | <50 | ND |
| 12/14/2004 | <24.1 | <24.1 | -- | |
| St. Joseph Creek at Beecher Rd. | 10/22/2004 | <13.4 | <13.4 | -- |
| 11/4/2004 | <21.1 | <21.1 | -- | |
| 11/17/2004 | <17.2 | <17.2 | -- | |
| 12/14/2004 | 21.8 | 21.8 | ND | |
| Main Branch River Raisin at Deerfield Rd | 12/6/2004 | <44 | <44 | -- |
| 1/5/2005 | 60.9 | 40.6 | ND | |
| Main Branch River Raisin at Crockett Rd | 12/6/2004 | <9.050 | <9.050 | -- |
| 1/5/2005 | 18.0 | <18.0 | ND | |
| Adrian Water Works Raw Influent | 1/17/2005 | <15.4 | <15.4 | -- |
| 1/24/2005 | 28.4 | 14.2 | ND | |
| 1/31/2005 | 13.5 | <13.5 | ND | |
| 2/7/2005 | 75.8 | <25.2 | ND | |
| 2/14/2005 | <13.1 | <13.1 | -- | |
| Blissfield Water Works Raw Influent | 12/14/2004 | <24.1 | <24.1 | ND |
| 1/17/2005 | 50.6 | <12.6 | ND | |
| 1/24/2005 | <12.7 | <12.7 | -- | |
| 1/31/2005 | <19.4 | 19.4 | -- | |
| 2/7/2005 | <11.6 | <11.6 | -- | |
| 2/14/2005 | 64.3 | <16.1 | ND | |
| Deerfield Water WorksRaw Influent | 1/18/2005 | 15.7 | <15.7 | ND |
| 1/24/2005 | <19.0 | <19.0 | -- | |
| 1/31/2005 | <14.7 | <14.7 | -- | |
| 2/7/2005 | 39.6 | <13.2 | ND | |
| 2/14/2005 | 64.3 | <16.1 | ND | |
| Bear Creek 1 (at Medina Rd.) | 11/9/2004 | <8.8 | 8.8 | -- |
| Wolf Creek at Forrister Rd. | 10/22/2004 | <20 | <20 | -- |
| 11/4/20041 | <13.1 | <13.1 | -- | |
| 1/17/2004 | <10.3 | <10.3 | -- | |
| Bear Creek 2 (at Morse Rd.) | 11/9/2004 | <18.2 | <18.2 | -- |
3.1.2. Genotyping of Cryptosporidium in Rural Water Samples
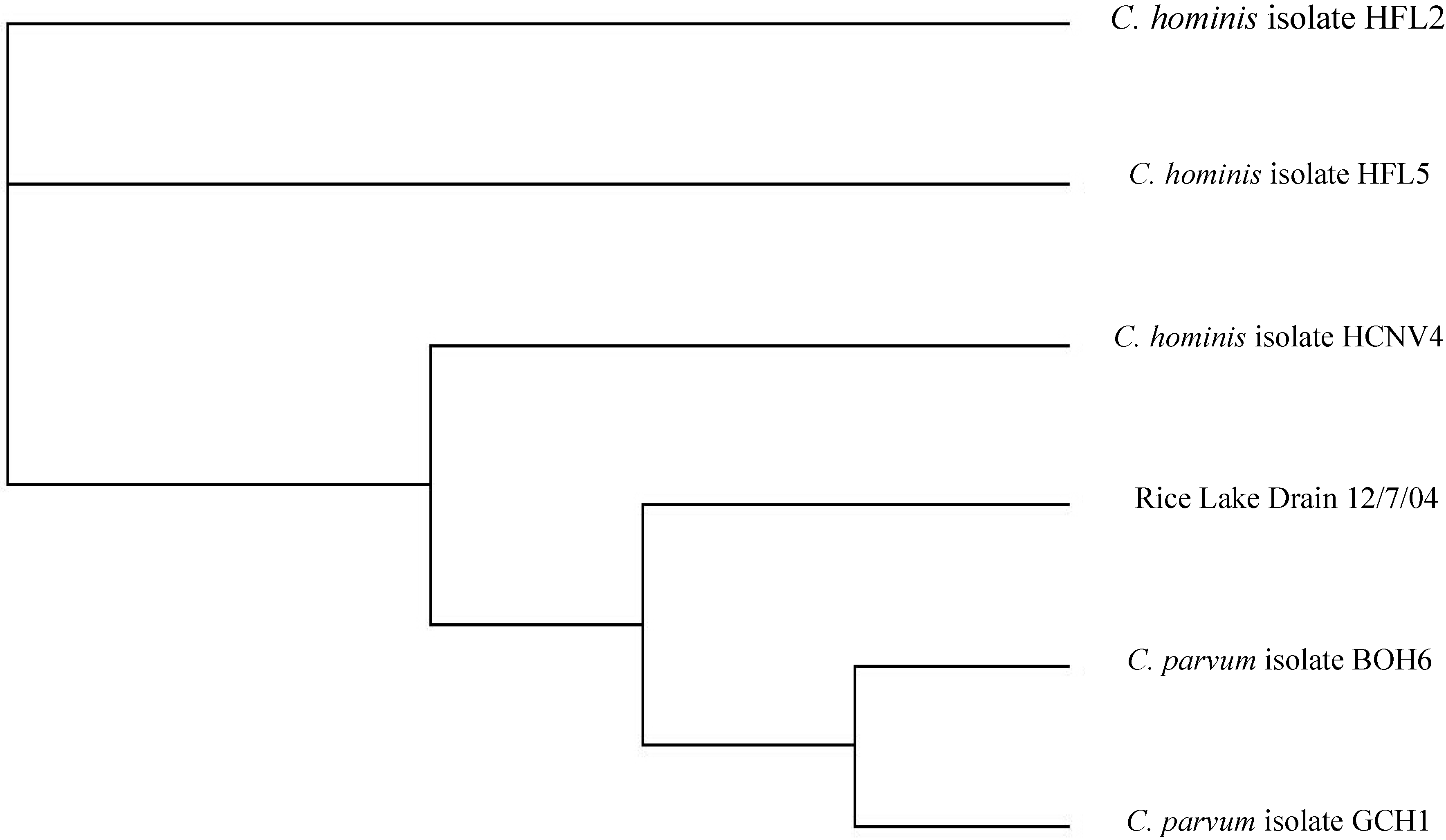
3.2. Relationship between Disease and Land Use
| Variable | Cryptosporidiosis Total Cases | Giardiasis Total Cases | ||
|---|---|---|---|---|
| Urban | Rural | Urban | Rural | |
| Number of Disease Cases | 39 ** | 113 ** | 405 ** | 467 ** |
| Cases/Population | 1.3 × 10−4 | 2.4 × 10−4 | 1.3 × 10−3 | 9.8 × 10−4 |
| Cases/Area | 0.19 ** | 0.03 ** | 1.9 ** | 0.10 ** |
| Cases/Population Density | 0.03 | 1.1 | 0.28 | 4.5 |
| Factor | Total Giardiasis Cases | Total Cryptosporidiosis Cases | % Urban Area | % Rural Area |
|---|---|---|---|---|
| % Urban Area | 0.79 | 0.60 | - | - |
| % Rural Area | −0.79 | −0.59 | - | - |
| Population | 0.92 | 0.79 | 0.75 | −0.75 |
| Population Density | 0.80 | 0.61 | 0.98 | −0.98 |
| % White | −0.69 | −0.55 | −0.78 | 0.76 |
| Median Age | −0.64 | −0.49 | −0.55 | 0.56 |
| Mean Travel Time to Work | −0.75 | −0.66 | −0.65 | 0.65 |
3.3. Lessons Learned: Implications for Management

4. Conclusions
Supplementary Files
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- Baldursson, S.; Karanis, P. Waterborne transmission of protozoan parasites: Review of worldwide outbreaks—An update 2004–2010. Water Res. 2011, 45, 6603–6614. [Google Scholar] [CrossRef]
- Fletcher, S.M.; Stark, D.; Harkness, J.; Ellis, J. Enteric protozoa in the developed world: A public health perspective. Clin. Microbiol. Rev. 2012, 25, 420–449. [Google Scholar] [CrossRef]
- Jellison, K.L.; Hemond, H.F.; Schauer, D.B. Sources and species of Cryptosporidium oocysts in the Wachusett Reservoir watershed. Appl. Environ. Microbiol. 2002, 68, 569–575. [Google Scholar] [CrossRef]
- Feng, Y.; Alderisio, K.A.; Yang, W.; Blancero, L.A.; Kuhne, W.G.; Nadareski, C.A.; Reid, M.; Xiao, L. Cryptosporidium genotypes in wildlife from a New York watershed. Appl. Environ. Microbiol. 2007, 73, 6475–6483. [Google Scholar] [CrossRef]
- Hunter, P.R.; Thompson, R.C.A. The zoonotic transmission of Giardia and Cryptosporidium. Int. J. Parasitol. 2005, 35, 1181–1190. [Google Scholar] [CrossRef]
- Wilkes, G.; Ruecker, N.J.; Neumann, N.F.; Gannon, V.P.J.; Jokinen, C.; Sunohara, M.; Topp, E.; Pintar, K.D.M.; Edge, T.A.; Lapen, D.R. Spatiotemporal analysis of Cryptosporidium species/genotypes and relationships with other zoonotic pathogens in surface water from mixed-use watersheds. Appl. Environ. Microbiol. 2013, 79, 434–448. [Google Scholar] [CrossRef]
- Fuchslin, H.P.; Kotzch, S.; Egli, T. Cryptosporidium spp. in drinking water. Samples from rural sites in Switzerland. Swiss Med. Wkly. 2012, 142. [Google Scholar] [CrossRef]
- Castro-Hermida, J.A.; Garcia-Presedo, I.; Almeida, A.; Gonzalez-Warleta, M.; Correia Da Costa, J.M.; Mezo, M. Presence of Cryptosporidium spp. and Giardia duodenalis through drinking water. Sci. Total Environ. 2008, 405, 45–53. [Google Scholar] [CrossRef]
- Galvan, A.L.; Magnet, A.; Izquierdo, F.; Vadillo, C.F.; Peralta, R.H.; Angulo, S.; Fenoy, S.; del Aguila, C. A year-long study of Cryptosporidium species and subtypes in recreational, drinking and wastewater from the central area of Spain. Sci. Total Environ. 2014, 468, 368–375. [Google Scholar] [CrossRef]
- Plutzer, J.; Tako, M.H.; Marialigeti, K.; Torokne, A.; Karanis, P. First investigations into the prevalence of Cryptosporidium and Giardia spp. in Hungarian drinking water. J. Water Health 2007, 5, 573–584. [Google Scholar] [CrossRef]
- Feng, Y.; Zhao, X.; Chen, J.; Jin, W.; Zhou, X.; Li, N.; Wang, L.; Xiao, L. Occurrence, source, and human infection potential of Cryptosporidium and Giardia spp. in source and tap water in Shanghai, China. Appl. Environ. Microbiol. 2011, 77, 3069–3616. [Google Scholar]
- Mons, C.; Dumetre, A.; Gosselin, S.; Galliot, C.; Moulin, L. Monitoring of Cryptosporidium and Giardia river contamination in Paris area. Water Res. 2009, 43, 211–217. [Google Scholar]
- Pollock, K.G.J.; Ternent, H.E.; Mellor, D.J.; Chalmers, R.M.; Smith, H.V.; Ramsay, C.N.; Innocent, G.T. Spatial and temporal epidemiology of sporadic human cryptosporidiosis in Scotland. Zoonoses Public Health 2010, 57, 487–492. [Google Scholar] [CrossRef]
- Lake, I.R.; Nichols, G.; Harrison, F.C.D.; Bentham, G.; Sari Kovats, R.; Grundy, C.; Hunter, P.R. Using infectious intestinal disease surveillance data to explore illness aetiology; A cryptosporidiosis case study. Health Place 2009, 15, 333–339. [Google Scholar] [CrossRef]
- Odoi, A.; Martin, S.W.; Michel, P.; Middleton, D.; Holt, J.; Wilson, J. Investigation of clusters of giardiasis using GIS and a spatial scan statistic. Int. J. Health Geogr. 2004, 3. [Google Scholar] [CrossRef] [Green Version]
- US Census Bureau US Census Bureau. Available online: www.census.gov/ (accessed on 10 July 2014).
- Donovan, M.L.; Nesslage, G.M.; Skillen, J.J.; Maurer, B.A. The Michigan Gap Analysis Project Final Report 2004. Available online: www.fw.msu.edu/~maurerb/gap/report.html (accessed on 19 September 2014).
- Michigan Department of Community Health Michigan Rural Health Profile: A Report on the Health Trends and Resources of Rural Michigan 1990–2005. Available online: www.michigan.gov/documents/mdch/MichiganRuralHealthProfile-2008-0801_243955_7.pdf (accessed on 19 September 2014).
- Michigan Department of Environmental Quality DEQ—Drinking Water. Available online: www.michigan.gov/deq/0,4561,7-135-3313_3675---,00.html (accessed on 10 July 2014).
- US EPA, Office of Water. Safe Drinking Water Information System. Available online: http://water.epa.gov/scitech/datait/databases/drink/sdwisfed/index.cfm (accessed on 10 July 2014).
- Michigan Department of Community Health Reportable Infections Diseases in Michigan 2003–2007 Summary 2008. Available online: www.michigan.gov/documents/mdch/Final_Reportable_ID_2003_to_2007_Summary_251984_7.pdf (accessed on 19 September 2014).
- Yoder, J.S.; Wallace, R.M.; Collier, S.A.; Beach, M.J.; Hlavsa, M.C. Cryptosporidiosis surveillance—United States, 2009–2010. MMWR Surveill. Summ. 2012, 61, 1–12. [Google Scholar]
- Furness, B.W.; Beach, M.J.; Roberts, J.M. Giardiasis Surveillance—United States, 1992–1997. MMWR Surveill. Summ. 2000, 49, 1–13. [Google Scholar]
- Hlavsa, M.C.; Watson, J.C.; Beach, M.J. Cryptosporidiosis surveillance—United States 1999–2002. MMWR Surveill. Summ. 2005, 54, 1–8. [Google Scholar]
- US EPA. LT2 Rule. Available online: http://water.epa.gov/lawsregs/rulesregs/sdwa/lt2/regulations.cfm#lt2data (accessed on 10 July 2014).
- US EPA. Method 1622: Cryptosporidium in Water by Filtration/IMS/FA. 2005; EPA 815-R-05-001. Available online: http://nepis.epa.gov/Adobe/PDF/P100997Q.PDF (accessed on 10 July 2014). [Google Scholar]
- US EPA. Method 1623: Cryptosporidium and Giardia in Water by Filtration/IMS/FA. 2005; EPA 815-R-05-002. Available online: http://water.epa.gov/scitech/methods/cwa/bioindicators/upload/method_1623.pdf (accessed on 10 July 2014). [Google Scholar]
- US EPA, Region 5. River Raisin. Available online: http://epa.gov/greatlakes/aoc/river-raisin/index.html (accessed on 28 July 2014).
- Singh, S. Investigation of Bacterial Fecal Indicators and Coliphage Virus in Sediment and Surface Water of Parks and Beaches along the Grand River (Michigan) and Lake Michigan (Michigan); Michigan State University: East Lansing, MI, USA, 2007. [Google Scholar]
- Ives, R.L. Giardiasis and Cryptosporidiosis in the Urban-Rural Spectrum; Michigan State University: East Lansing, MI, USA, 2011. [Google Scholar]
- Wilczynski, J.A.; Ives, R.; Peters, S.; Henderson, T.; House, J.; Hill, V.; Schneeberger, C.; Xiao, L.; Dearen, T.; Webeck, J. Outbreak of cyptosporidiosis associated with a firefighting response—Indiana and Michigan, June 2011. Morb. Mortal. Wkly. Rep. 2012, 61, 153–156. [Google Scholar]
- Slifko, T.R.; Huffman, D.E.; Rose, J.B. A most-probable-number assay for enumeration of infectious Cryptosporidium parvum oocysts. Appl. Environ. Microbiol. 1999, 65, 3936–3941. [Google Scholar]
- Xiao, L.H.; Escalante, L.; Yang, C.F.; Sulaiman, I.; Escalante, A.A.; Montali, R.J.; Fayer, R.; Lal, A.A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl. Environ. Microbiol. 1999, 65, 1578–1583. [Google Scholar]
- Xiao, L.H.; Alderisio, K.; Limor, J.; Royer, M.; Lal, A.A. Identification of species and sources of Cryptosporidium oocysts in storm waters with a small-subunit rRNA-based diagnostic and genotyping tool. Appl. Environ. Microbiol. 2000, 66, 5492–5498. [Google Scholar]
- Sulaiman, I.M.; Xiao, L.H.; Yang, C.F.; Escalante, L.; Moore, A.; Beard, C.B.; Arrowood, M.J.; Lal, A.A. Differentiating human from animal isolates of Cryptosporidium parvum. Emerg. Infect. Dis. 1998, 4, 681–685. [Google Scholar] [CrossRef]
- Peng, M.M.; Xiao, L.H.; Freeman, A.R.; Arrowood, M.J.; Escalante, A.A.; Weltman, A.C.; Ong, C.S.L.; MacKenzie, W.R.; Lal, A.A.; Beard, C.B. Genetic polymorphism among Cryptosporidium parvum isolates: Evidence of two distinct human transmission cycles. Emerg. Infect. Dis. 1997, 3, 567–573. [Google Scholar] [CrossRef]
- Lindsay, D.S.; Upton, S.J.; Owens, D.S.; Morgan, U.M.; Mead, J.R.; Blagburn, B.L. Cryptosporidium andersoni n. sp (Apicomplexa : Cryptosporiidae) from cattle Bos taurus. J. Eukaryot. Microbiol. 2000, 47, 91–95. [Google Scholar]
- Peng, M.M.; Wilson, M.L.; Holland, R.E.; Meshnick, S.R.; Lal, A.A.; Xiao, L. Genetic diversity of Cryptosporidium spp. in cattle in Michigan: implications for understanding the transmission dynamics. Parasitol. Res. 2003, 90, 175–180. [Google Scholar] [CrossRef]
- Smerdon, W.J.; Nichols, T.; Chalmers, R.M.; Heine, H.; Reacher, M.H. Foot and mouth disease in livestock and reduced cryptosporidiosis in humans, England and Wales. Emerg. Infect. Dis. 2003, 9, 22–28. [Google Scholar] [CrossRef]
- Thompson, R.C.A.; Reynoldson, J.A.; Lymbery, A.J. Giardia: from Molecules to Disease; CAB International: Wallingford, UK, 1994. [Google Scholar]
- Lake, I.R.; Harrison, F.C.D.; Chalmers, R.M.; Bentham, G.; Nichols, G.; Hunter, P.R.; Kovats, R.S.; Grundy, C. Case-control study of environmental and social factors influencing cryptosporidiosis. Eur. J. Epidemiol. 2007, 22, 805–811. [Google Scholar] [CrossRef]
- Ongerth, J.E. LT2 Cryptosporidium data: What do they tell us about Cryptosporidium in surface water in the United States? Environ. Sci. Technol. 2013, 47, 4029–4038. [Google Scholar] [CrossRef]
- Austin, Z.; Alcock, R.E.; Christley, R.M.; Haygarth, P.M.; Heathwaite, A.L.; Latham, S.M.; Mort, M.; Oliver, D.M.; Pickup, R.; Wastling, J.M.; Wynne, B. Policy, practice and decision making for zoonotic disease management: Water and Cryptosporidium. Environ. Int. 2012, 40, 70–78. [Google Scholar] [CrossRef]
- Van Herk, F.H.; McAllister, T.A.; Cockwill, C.L.; Guselle, N.; Larney, F.J.; Miller, J.J.; Olson, M.E. Inactivation of Giardia cysts and Cryptosporidium oocysts in beef feedlot manure by thermophilic windrow composting. Compost Sci. Util. 2004, 12, 235–241. [Google Scholar]
- Robertson, L.J.; Campbell, A.T.; Smith, H.V. Survival of Cryptosporidium parvum oocysts under various environmental pressures. Appl. Environ. Microbiol. 1992, 58, 3494–3500. [Google Scholar]
- Zintl, A.; Keogh, B.; Ezzaty-Mirhashemi, M.; De Waal, T.; Scholz, D.; Mulcahy, G. Survival of Cryptosporidium parvum oocysts in the presence of hydrated lime. Vet. Rec. 2010, 166, 297–300. [Google Scholar] [CrossRef]
- Borchardt, M.A.; Chyou, P.H.; DeVries, E.O.; Belongia, E.A. Septic system density and infectious diarrhea in a defined population of children. Environ. Health Perspect. 2003, 111, 742–748. [Google Scholar] [CrossRef]
- Michigan Department of Environmental Quality Michigan BeachGuard System—Home. Available online: www.deq.state.mi.us/beach/Default.aspx (accessed on 15 July 2014).
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dreelin, E.A.; Ives, R.L.; Molloy, S.; Rose, J.B. Cryptosporidium and Giardia in Surface Water: A Case Study from Michigan, USA to Inform Management of Rural Water Systems. Int. J. Environ. Res. Public Health 2014, 11, 10480-10503. https://doi.org/10.3390/ijerph111010480
Dreelin EA, Ives RL, Molloy S, Rose JB. Cryptosporidium and Giardia in Surface Water: A Case Study from Michigan, USA to Inform Management of Rural Water Systems. International Journal of Environmental Research and Public Health. 2014; 11(10):10480-10503. https://doi.org/10.3390/ijerph111010480
Chicago/Turabian StyleDreelin, Erin A., Rebecca L. Ives, Stephanie Molloy, and Joan B. Rose. 2014. "Cryptosporidium and Giardia in Surface Water: A Case Study from Michigan, USA to Inform Management of Rural Water Systems" International Journal of Environmental Research and Public Health 11, no. 10: 10480-10503. https://doi.org/10.3390/ijerph111010480
APA StyleDreelin, E. A., Ives, R. L., Molloy, S., & Rose, J. B. (2014). Cryptosporidium and Giardia in Surface Water: A Case Study from Michigan, USA to Inform Management of Rural Water Systems. International Journal of Environmental Research and Public Health, 11(10), 10480-10503. https://doi.org/10.3390/ijerph111010480




