The Impact of Microbial Ecology and Chemical Profile on the Enhanced Biological Phosphorus Removal (EBPR) Process: A Case Study of Northern Wastewater Treatment Works, Johannesburg
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collection
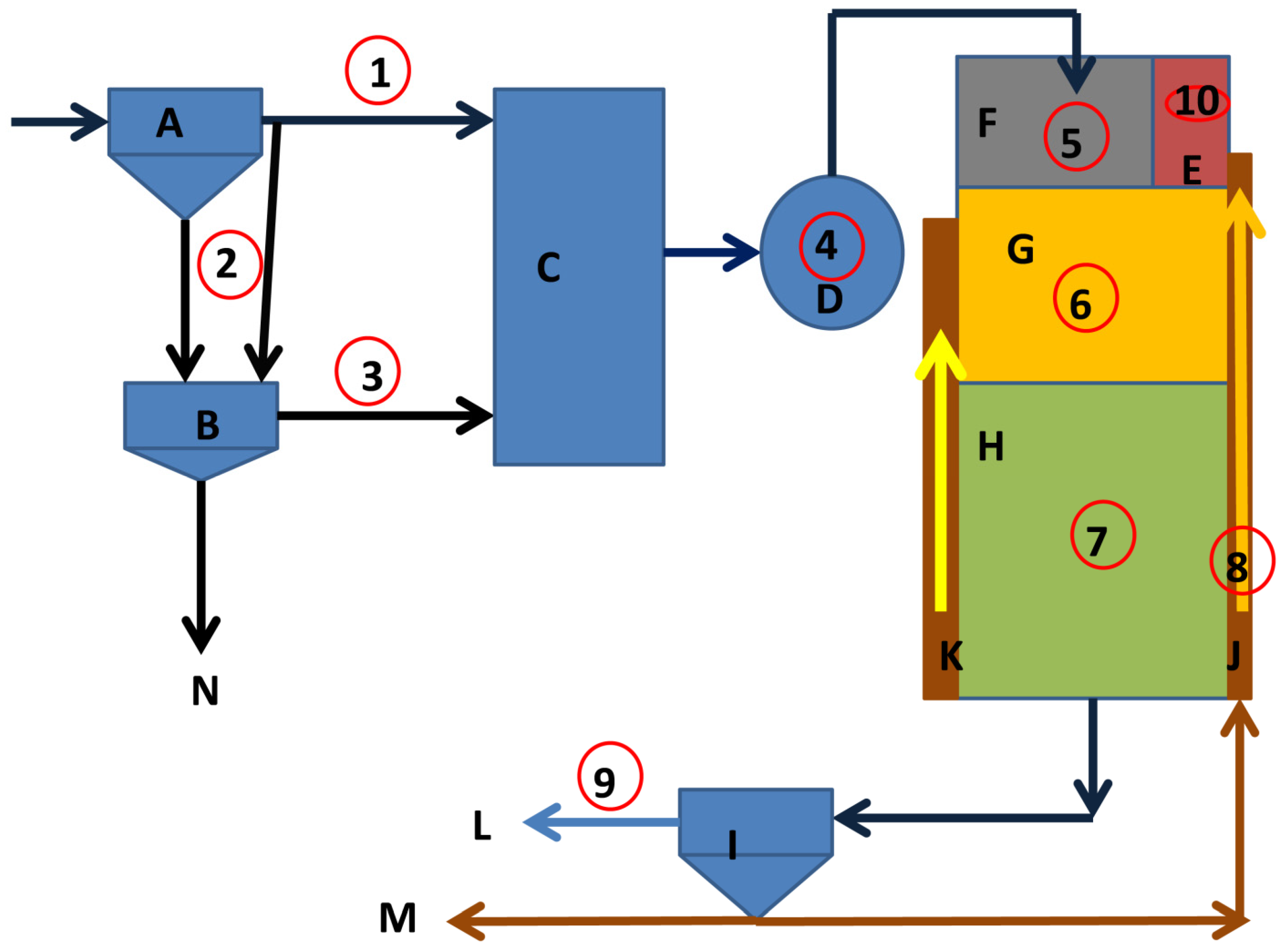
2.2. Chemical Reagents
2.3. Physicochemical Profile of the EBPR
2.4. Microbial Ecology
2.4.1. Isolation of DNA of the Microbial Species
2.4.2. PCR Amplification
| Name | Sequence | Reference |
|---|---|---|
| 27F | 5’AGRGTTTGATCMTGGCTCAG3’ | Tekere et al. [22] |
| 1492R | 5’GGTTACCTTGTTACGACTT3’ | |
| A1.4 | 5’ CGTATCGCCTCCCTCGCGCCATCA | |
| tctctatgcgAGRGTTTGATCMTGGCTCAG3’ | ||
| B1 | 5’CTATGCGCCTTGCCAGCCCGCTCAG | |
| GTATTACCGCGGCTGCTG3’ |
| Sequence | Variable Region (V 1-3) to Be Amplified | ||
|---|---|---|---|
| Elutriation tanks | Anaerobic zone | Aerobic zone | |
| Number of sequences | 2,417 | 2,436 | 2,098 |
| Total length of sequences (bp) | 617,990 | 671,243 | 579,638 |
| Average length of sequences (bp) | 200 | 120 | 180 |
2.4.3. Statistical Analyses
3. Results and Discussion
3.1. Physicochemical Parameters


| Elutriation Tank Unit-5 | Elutriation Tank Unit-3 | Elutriation Tank Unit-5 | Elutriation Tank Unit-3 | |
|---|---|---|---|---|
| 2012/04/12 | 2012/07/12 | |||
| VFA | Conc. (mg/L) | Conc. (mg/L) | Conc. (mg/L) | Conc. (mg/L) |
| Hexanoic acid, methyl ester | ND | ND | ND | ND |
| Heptanoic acid, methyl ester | ND | ND | ND | ND |
| Butanoic acid, 3-methyl- | ND | 17.38 ± 0.87 | 67.41 ± 3.37 | 56.34±2.82 |
| Pentanoic acid, 4-methyl- | ND | 13.64 ± 0.68 | ND | ND |
| Acetic acid | 57.65 ± 2.88 | 26.32 ± 1.32 | 58.85 ± 2.94 | 54.98 ± 2.75 |
| Propanoic acid | 38.61 ± 1.93 | 17.19 ± 0.86 | 44.45 ± 2.22 | 43.56 ± 2.18 |
| Isobutyric acid | 57.27 ± 2.86 | 29.67 ± 1.48 | 56.39 ± 2.82 | 43.96 ± 2.20 |
| Butanoic acid | 73.13 ± 3.66 | 49.68 ± 2.48 | 66.08 ± 3.30 | 50.67 ± 2.53 |
| Pentanoic acid | 76.6 ± 3.83 | 87.6 ± 4.38 | 69.45 ± 3.47 | 61.58 ± 3.08 |
| Heptanoic acid | 88.52 ± 4.43 | 33.98 ± 1.70 | 89.82 ± 4.49 | 53.97 ± 2.70 |
| 2012/05/14 | 2012/08/13 | |||
| Hexanoic acid, methyl ester | ND | ND | ND | ND |
| Heptanoic acid, methyl ester | 21.63 ± 1.08 | ND | ND | ND |
| Butanoic acid, 3-methyl- | 67.41 ± 3.37 | 37.95 ± 1.90 | 67.41±3.37 | 53.69±2.68 |
| Pentanoic acid, 4-methyl- | 80.15 ± 4.01 | 38.23 ± 1.91 | ND | ND |
| Acetic acid | 58.85 ± 2.94 | 39.68 ± 1.98 | 54.05±2.70 | 41.29±2.06 |
| Propanoic acid | 43.71 ± 2.19 | 43.32 ± 2.17 | 49.63±2.48 | 34.67±1.73 |
| Isobutyric acid | 57.27 ± 2.89 | 23.14 ± 1.16 | 56.39±2.82 | 43.95±2.20 |
| Butanoic acid | 65.2 ± 3.26 | 16.52 ± 0.83 | 70.49±3.52 | 61.84±3.09 |
| Pentanoic acid | 69.45 ± 3.47 | 12.65 ± 0.63 | 77.62±3.88 | 81.39±4.07 |
| Heptanoic acid | 92.39 ± 4.62 | 39.37 ± 1.97 | ND | 3.96± 0.20 |
| 2012/06/12 | ||||
| Hexanoic acid, methyl ester | ND | ND | ||
| Heptanoic acid, methyl ester | ND | ND | ||
| Butanoic acid, 3-methyl- | 67.41±3.37 | 43.39±2.17 | ||
| Pentanoic acid, 4-methyl- | ND | ND | ||
| Acetic acid | 57.65±2.88 | 45.25±2.26 | ||
| Propanoic acid | 43.71±2.19 | 36.54±1.83 | ||
| Isobutyric acid | 55.51±2.78 | 21.58±1.08 | ||
| Butanoic acid | 65.2±3.16 | 23.95±1.20 | ||
| Pentanoic acid | 69.45±3.26 | 46.39±2.32 | ||
| Heptanoic acid | 91.13±4.56 | 85.34±4.27 | ||



3.2. Microbial Ecology




4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kamika, I.; Momba, M.N.B. Comparing the tolerance limits of selected bacterial and protozoan species to nickel in wastewater systems. Sci. Total Environ. 2011, 410, 172–181. [Google Scholar] [CrossRef]
- Jern, W.N.G. Industrial Wastewater Treatment; Imperial College Press: Toh Tuck, Singapore, 2006; pp. 1–11. [Google Scholar]
- Kamika, I.; Momba, M.N.B. Assessing the resistance and bioremediation ability of selected bacterial and protozoan species to heavy metals in metal-rich industrial wastewater. BMC Microbiol. 2013, 13. [Google Scholar] [CrossRef]
- Van Vuuren, L. The state of water in South Africa—Are we heading for a crisis? Water Wheel 2009, 8, 31–33. [Google Scholar]
- Momba, M.N.B.; Sibewu, M. Survival of somatic and F-RNA coliphages in treated wastewater effluents and their impact on viral quality of the receiving water bodies in the Eastern Cape Province. J. Biol. Sci. 2009, 9, 648–654. [Google Scholar] [CrossRef]
- Seviour, R.J.; Mino, T.; Onuki, M. The microbiology of biological phosphorus removal in activated sludge systems. FEMS Microbiol. Rev. 2003, 27, 99–127. [Google Scholar] [CrossRef]
- Korstee, G.J.J.; Appeldoorn, K.J.; Bonting, C.F.C.; van Niel, E.W.J.; van Veen, H.W. Biology of polyphosphate-accumulating bacteria involved in enhanced biological phosphorus removal. FEMS Microbiol. Rev. 1994, 15, 137–153. [Google Scholar] [CrossRef]
- Lopez-Vazquez, C.M. The Competition between Polyphosphate-Accumulating Organisms and Glycogen-Accumulating Organisms: Temperature Effects and Modelling; country CRC Press: Balkema, South Africa, 2009; pp. 1–98. [Google Scholar]
- Akpor, O.B.; Momba, M.N.B.; Okonkwo, J.O. The effects of pH and temperature on phosphate and nitrate uptake by wastewater protozoa. Afr. J. Biotechnol. 2008, 7, 2221–2226. [Google Scholar]
- Mino, T.; van Loosdrecht, M.C.M.; Heijnen, J.J. Microbiology and biochemistry of the enhanced biological phosphorus removal process. Water Res. 1998, 32, 3193–3207. [Google Scholar] [CrossRef]
- Oehmen, A.; Lemos, P.C.; Carvalho, G.; Yuan, Z.; Keller, J.; Blackall, L.L.; Reis, M.A.M. Advances in enhanced biological phosphorus removal: From micro to macro scale. Water Res. 2007, 41, 2271–2300. [Google Scholar] [CrossRef]
- He, S.; Bishop, F.I.; McMahon, K.D. Bacterial community and “Candidatus accumulibaster” population in laboratory-scale enhanced biological phosphate removal reactors. Appl. Environ. Microbiol. 2010, 76, 5479–5487. [Google Scholar] [CrossRef]
- Henze, M.; van Loosdrecht, M.C.M.; Ekama, G.; Brdjanovic, D. Biological Wastewater Treatment: Principles, Modelling and Design; IWA Publishing: London, UK, 2008; pp. 33–220. [Google Scholar]
- Oehmen, A.; Zeng, R.J.; Saunders, A.M.; Blackall, L.L.; Keller, J.; Yuan, Z. Anaerobic and aerobic metabolism of glycogen-accumulating organisms selected with propionate as the sole carbon source. Microbiology 2006, 152, 2767–2778. [Google Scholar] [CrossRef]
- Pijuan, M.; Ye, L.; Yuan, Z. Free nitrous acid inhibition on the aerobic metabolism of poly-phosphate accumulating organisms. Water Res. 2010, 44, 6063–6072. [Google Scholar] [CrossRef]
- Momba, M.N.B.; Cloete, T.E. The relationship of biomass to phosphate uptake by Acinetobacter junii in activated sludge mixed liquor. Water Res. 1996, 30, 364–370. [Google Scholar] [CrossRef]
- Momba, M.N.B.; Cloete, T.E. Biomass relationship to growth and phosphate uptake of Pseudomonas fluorescens, Escherichia coli and Acinetobacter radioresistens in mixed liquor medium. J. Ind. Microbiol. 1996, 16, 364–369. [Google Scholar] [CrossRef]
- Crocetti, G.R.; Banfield, J.F.; Keller, J.; Bond, P.L.; Blackall, L.L. Glycogen accumulating organisms in laboratory-scale and full-scale wastewater treatment processes. Microbiology 2002, 148, 3353–3364. [Google Scholar]
- Johannesburg’s Water Cycle. Available online: http:/www.eco-h2o.co.za/2010/04/12/johannesburgs-water-cycle (accessed on 17 December 2012).
- American Public Health Association (APHA). Standard Methods for the Examination of Water and Wastewater, 20th ed.; APHA: Washington, DC, USA, 2001. [Google Scholar]
- Ozutsumi, Y.; Tajima, K.; Takenaka, A.; Itabashi, H. The effect of protozoa on the composition of rumen bacteria in cattle using 16S rRNA gene clone libraries. Biosci. Biotechnol. Biochem. 2005, 69, 499–506. [Google Scholar] [CrossRef]
- Tekere, M.; Lotter, A.; Olivier, J.; Jonker, N.; Venter, S. Metagenomic analysis of bacterial diversity of Siloam hot water spring, Limpopo, South Africa. Afr. J. Biotechnol. 2011, 10, 18005–18012. [Google Scholar]
- Water Research Commission (WRC). Manual for the Design and Operation of Nutrient Removal Plants; WRC: Pretoria, South Africa, 1984. [Google Scholar]
- Rossle, W.H.; Pretorius, W.A. A review of characterisation requirements for in-line prefermenters. Paper 1: Wastewater characterisation. Water SA 2001, 27, 405–412. [Google Scholar]
- Rossle, W.H.; Pretorius, W.A. A review of characterisation requirements for in-line prefermenters. Paper 2: Process characterisation. Water SA 2001, 27, 413–422. [Google Scholar]
- Bouzas, A.; Ribes, J.; Ferrer, J.; Seco, A. Fermentation and elutriation of primary sludge: Effect of SRT on process performance. Water Res. 2007, 41, 747–756. [Google Scholar] [CrossRef]
- Pisco, A.R.; Bengtsson, S.; Wrker, A.; Reis, M.A.; Lemos, P.C. Community structure evolution and enrichment of glycogen-accumulating organisms producing polyhydroxyalkanoates from fermented molasses. Appl. Environ. Microbiol. 2009, 75, 4676–4688. [Google Scholar] [CrossRef]
- Hill, D.T.; Holmberg, R.D. Long chain volatile fatty acid relationships in anaerobic digestion of swine waste. Biol. Waste 1988, 23, 195–214. [Google Scholar] [CrossRef]
- Hood, C.; Randall, A.A. A biochemical hypothesis explaining the response of enhanced biological phosphorus removal biomass to organic substrates. Water Res. 2011, 35, 2758–2766. [Google Scholar] [CrossRef]
- Canizares, P.; Rodriguez, L.; Villasenor, J.; Rodriguez, J. Effect of wastewater composition on the development of an activated sludge biological phosphorus removal system. Environ. Technol. 1999, 20, 159–169. [Google Scholar] [CrossRef]
- Tracy, K.D.; Flammino, A. Kinetics of Biological Phosphorus Removal. In Proceedings of the 58th Annual Water Pollution Control Federation Conference, Kansas City, MO, USA, 7–10 October 1985.
- Smolders, G.J.F.; van Der Meij, J.; van Loosdrecht, M.C.M.; Heijnen, J.J. Stoichiometric model of the aerobic metabolism of the biological phosphorus removal process. Biotechnol. Bioeng. 1994, 44, 837–848. [Google Scholar] [CrossRef]
- Liu, W.T.; Mino, T.; Nakamura, K.; Matsuo, T. Glycogen accumulating population and its anaerobic substrate uptake in anaerobic-aerobic activated sludge without biological phosphorus removal. Water Res. 1996, 30, 75–82. [Google Scholar] [CrossRef]
- Schuler, A.J.; Jenkins, D. Effects of pH on enhanced biological phosphorus removal metabolisms. Water Sci. Technol. 2002; 46, 171–178. [Google Scholar]
- United States Environmental Protection Agency (USEPA). National Section 303(d) List Fact Sheet; USEPA: Las Vegas, NV, USA. Available online: http://iaspub.epa.gov/waters/national_rept.control (accessed on 28 October 2013).
- Glass, C.; Silverstein, J.A.; Denton, L. Bacterial Populations in Activated Sludge Denitrifying High Nitrate Waste Reflect pH Differences. In Proceedings of the 2nd International Conference on Microorganisms in Activated Sludge and Biofilm Processes, Berkeley, CA, USA, 21–23 July 1997.
- Jenkins, D.; Tandoi, V. The applied microbiology of enhanced biological phosphate removal—Accomplishments and needs. Water Res. 1991, 25, 1471–1478. [Google Scholar]
- Cech, J.S.; Hartman, P. Competition between polyphosphate and polysaccharide accumulating bacteria in enhanced biological phosphate removal systems. Water Res. 1993, 27, 1219–1225. [Google Scholar]
- Liu, W.T.; Nielsen, A.T.; Wu, J.H.; Tsai, C.S.; Matsuo, Y.; Molin, S. In situ identification of polyphosphate- and polyhydroxyalkanoate-accumulating traits for microbial populations in a biological phosphorus removal process. Environ. Microbiol. 2001, 3, 110–122. [Google Scholar]
- Wagner, M.; Erhart, R.; Manz, W.; Amann, R.; Lemmer, H.; Wedi, D.; Schleifer, K.H. Development of a ribosomal-RNA-targetedoligonucleotide probe specific for the genus Acinetobacter and its application for in situ monitoring in activated sludge. Appl. Environ. Microbiol. 1994, 60, 2–800. [Google Scholar]
- Ren, Y.; Wei, C.H.; Xiao, K.J. Characterisation of microorganisms responsible for EBPR in a sequencing batch reactor by using the 16S rDNA-DGGE method. Water SA 2007, 33, 123–128. [Google Scholar]
- Heylen, K.; Vanparys, B.; Wittebolle, L.; Verstraete, W.; Boon, N.; de Vos, P. Cultivation of denitrifying bacteria: Optimization of isolation conditions and diversity study. Appl. Environ. Microbiol. 2006, 72, 2637–2643. [Google Scholar]
- Snaidr, J.; Amann, R.; Huber, I.; Ludwig, W.; Schleifer, K.H. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 1997, 63, 2884–2896. [Google Scholar]
- Mullan, A.; Quinn, J.P.; McGrath, J.W. Enhanced phosphate uptake and polyphosphate accumulation in Burkholderia cepacia grown under low-pH conditions. Microbial Ecol. 2002, 44, 69–77. [Google Scholar]
- Muszynski, A.; Lebkowska, M.; Tabernacka, A.; Mitobedzka, A. From macro to lab-scale: Changes in bacterial community led to deterioration of EBPR in lab reactor. Cent. Eur. J. Biol. 2013, 8, 130–142. [Google Scholar]
- Burow, L.C.; Kong, Y.; Nielsen, J.L.; Blackall, L.L.; Nielsen, P.H. Abundance and ecophysiology of Defluviicoccus spp., glycogen-accumulating organisms in full-scale wastewater treatment processes. Microbiology 2007, 153, 178–185. [Google Scholar]
- Beer, M.; Kong, Y.H.; Seviour, R.J. Are some putative glycogen accumulating organisms (GAO) in anaerobic: Aerobic activated sludge systems members of the α-Proteobacteria? Microbiology 2004, 150, 2267–2275. [Google Scholar]
- Schramm, A.; de Beer, D.; van den Heuvel, J.C.; Ottengraf, S.; Amann, R. Microscale distribution of populations and activities of Nitrosospira and Nitrospira spp. along a macroscale gradient in a nitrifying bioreactor: Quantification by in situ hybridisation and the use of microsensors. Appl. Environ. Microbiol. 1999, 65, 3690–3696. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kamika, I.; Coetzee, M.; Mamba, B.B.; Msagati, T.; Momba, M.N.B. The Impact of Microbial Ecology and Chemical Profile on the Enhanced Biological Phosphorus Removal (EBPR) Process: A Case Study of Northern Wastewater Treatment Works, Johannesburg. Int. J. Environ. Res. Public Health 2014, 11, 2876-2898. https://doi.org/10.3390/ijerph110302876
Kamika I, Coetzee M, Mamba BB, Msagati T, Momba MNB. The Impact of Microbial Ecology and Chemical Profile on the Enhanced Biological Phosphorus Removal (EBPR) Process: A Case Study of Northern Wastewater Treatment Works, Johannesburg. International Journal of Environmental Research and Public Health. 2014; 11(3):2876-2898. https://doi.org/10.3390/ijerph110302876
Chicago/Turabian StyleKamika, Ilunga, Martie Coetzee, Bhekie Brilliance Mamba, Titus Msagati, and Maggy N. B. Momba. 2014. "The Impact of Microbial Ecology and Chemical Profile on the Enhanced Biological Phosphorus Removal (EBPR) Process: A Case Study of Northern Wastewater Treatment Works, Johannesburg" International Journal of Environmental Research and Public Health 11, no. 3: 2876-2898. https://doi.org/10.3390/ijerph110302876





