Assessing the Groundwater Quality at a Saudi Arabian Agricultural Site and the Occurrence of Opportunistic Pathogens on Irrigated Food Produce
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling Site and Sampling Procedure

2.2. Measurement of Nutrient Content, Coliforms and Numbers of Total Cells in Groundwater
2.3. Groundwater Filtration and DNA Extraction
2.4. Quantitative PCR (qPCR)
2.5. 16S rRNA Gene Amplicon-based High-throughput Sequencing and Analysis
2.6. Isolation and Phylogenetic Identification of Bacterial Isolates Based on 16S rRNA Genes
2.7. Antibiotic Susceptibility of Pseudomonas Aeruginosa
2.8. Quantitative Microbial Risk Assessment (QMRA)
2.9. Nucleotide Sequence Accession Numbers
3. Results
3.1. Chemical and Microbial Quality of Groundwater Samples
| Well Name | Group Number | Total Nitrogen, TN | Non-Particulate Organic Carbon, NPOC | Total Coliforms | Fecal Coliforms | 16S rRNA Gene Copies |
|---|---|---|---|---|---|---|
| Average (mg/L) ± Standard Deviation | MPN/100 mL | Copies/L ± Standard Deviation | ||||
| A (I) | 1 | 19.6 ± 0.2 | 69.1 ± 2.0 | None detected | 1.66 × 109 ± 2.52 × 108 | |
| B (I) | 1 | 23.0 ± 0.1 | 16.1 ± 0.2 | 1.97 × 107 ± 5.87 × 105 | ||
| C (I) | 2 | 53.7 ± 0.3 | 70.6 ± 2.8 | 5.19 × 107 ± 4.96 × 106 | ||
| A (II) | 1 | 23.1 ± 0.6 | 33.6 ± 0.3 | 5.46 × 107 ± 1.25 × 106 | ||
| B (II) | 1 | 37.6 ± 1.0 | 34.9 ± 14.2 | 2.32 × 107 ± 9.14 × 105 | ||
| C (II) | 2 | 61.3 ± 5.6 | 22.7 ± 2.2 | 3.94 × 107 ± 3.63 × 106 | ||
| D | 2 | 55.1 ± 2.9 | 10.6 ± 0.9 | >1600 | 12 | 1.28 × 1010 ± 2.56 × 108 |
| E | 2 | 40.8 ± 0.5 | 12.1 ± 0.01 | >1600 | 1600 | 1.14 × 109 ± 5.94 × 108 |
| F | 2 | 49.5 ± 0.6 | 68.8 ± 0.5 | >1600 | 920 | 4.53 × 107 ± 2.65 × 105 |
| G | 2 | 42.2 ± 2.2 | 69.9 ± 2.9 | None detected | 2.95 × 107 ± 2.49 × 105 | |
| H | 1 | 15.2 ± 0.1 | 70.6 ± 0.3 | 3.14 × 106 ± 5.78 × 105 | ||
3.2. qPCR-based Fecal Source Tracking
3.3. Multivariate Analysis of Microbial Communities on a Metric Multidimensional Scaling Plot
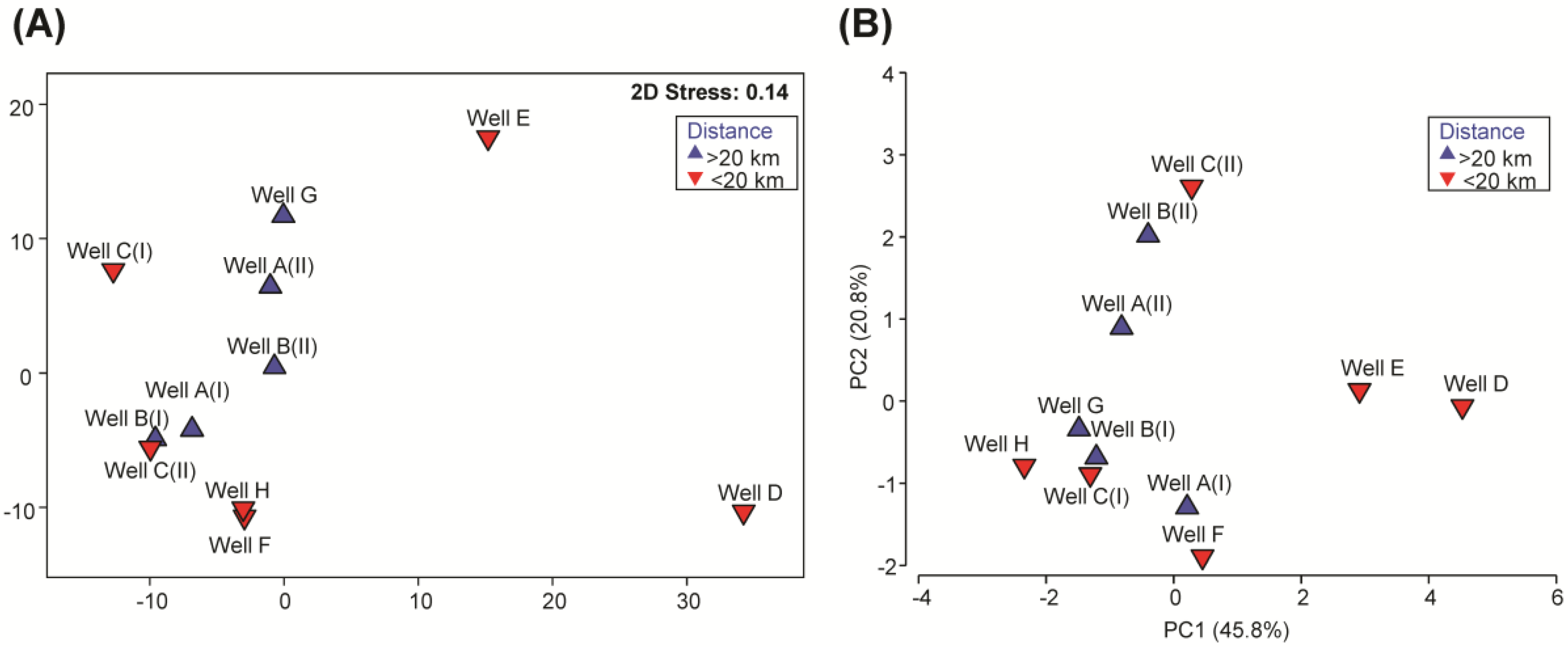
3.4. Multivariate Analysis of Water Quality Parameters and Correlation to the Microbial Pattern
3.5. Molecular-based Detection of Genera Associated with Opportunistic Pathogens and Bacterial Isolation from Groundwater Samples

3.6. Bacterial Isolation in Irrigated Fruits
3.7. Microbial Risk Associated with Ingestion of Tomatoes and Peppers Harvested from Sampling Sites
| Parameters | Annotation | Assumed Value | Reference |
|---|---|---|---|
| Average weight per person (kg) | A | 70 | [27] |
| Total amount of fruits consumed (g/kg/d, assuming that it is an equal portion of only tomatoes and green pepper) | B | 2.9 | |
| Proportion of consumed fruits amounting from the peels | C | 0.10 | |
| Transmission probability of bacterium from water to fruit surfaces | D | 2.00 × 10−6 | [26] |
| Median cell numbers of genus Pseudomonas per 50 g of peels over 95% confidence interval | E | 1.73 × 108 | |
| Median cell numbers of genus Enterococcus per 50 g of peels over 95% confidence interval | F | 9.20 × 108 | |
| Exposure dose of P. aeruginosa (cells/event) = A × B × C × D × E / 50 g | 1.40 × 102 | ||
| k of P. aeruginosa (Exponential model; derived from the LD50 dose of P. aeruginosa required for infections via murine gastrointestinal tract) | 1.87 × 10−8 | [29] | |
| Point estimate of risk arising from P. aeruginosa = 1 − exp (−k × exposure dose) | 2.62 × 10−6 | ||
| Annual risk arising from P. aeruginosa = 1 − (1-point estimate)^365 days per year | 9.55 × 10−4 | ||
| Exposure dose of E. faecalis (cells/event) = A × B × C × D × F / 50 g | 7.47 × 102 | ||
| k of E. faecalis (Exponential model; derived from LD50 dose of E. faecalis required for peritonitis via blood injection) | 2.19 × 10−11 | [28] | |
| Point estimate of risk arising from E. faecalis = 1 − exp (−k × exposure dose) | 1.64 × 10−8 | ||
| Annual risk arising from E. faecalis = 1 − (1-point estimate)^365 days per year | 5.98 × 10−6 | ||
4. Discussion
5. Conclusions
Supplementary Files
Supplementary File 1Acknowledgement
Author Contributions
Conflicts of Interest
References
- Dabbagh, A.E.; Abderrahman, W.A. Management of groundwater resources under various irrigation water use scenarios in Saudi Arabia. Arabian J. Sci. Eng. 1997, 22, 47–64. [Google Scholar]
- Zaharani, K.H.; Al-Shayaa, M.S.; Baig, M.B. Water conservation in the Kingdom of Saudi Arabia for better environment: Implications for extension and education. Bulg. J. Agric. Sci. 2011, 17, 389–395. [Google Scholar]
- International Water Management Institute (IWMI). Does Food Trade Save Water? The Potential Role of Food Trade in Water Scarcity Mitigation; International Water Management Institute: Colombo, Sri Lanka, 2007. [Google Scholar]
- Hoekstra, A.Y.; Chapagain, A.K. Water footprints of nations: Water use by people as a function of their consumption pattern. Water Resour. Manag. 2007, 21, 35–48. [Google Scholar] [CrossRef]
- Al-Mazrou, Y.Y. Food poisoning in Saudi Arabia. Potential for prevention? Saudi Med. J. 2004, 25, 11–14. [Google Scholar] [PubMed]
- Al-Ghamdi, M.A.; Bentham, G.; Hunter, P.R. Environmental risk factors for diarrhoea among male schoolchildren in Jeddah city, Saudi Arabia. J. Water Health. 2009, 7, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Cheong, S.; Lee, C.; Song, S.W.; Choi, W.C.; Lee, C.H.; Kim, S.J. Enteric viruses in raw vegetables and groundwater used for irrigation in South Korea. Appl. Environ. Microb. 2009, 75, 7745–7751. [Google Scholar] [CrossRef] [PubMed]
- Steele, M.; Odumeru, J. Irrigation water as source of foodborne pathogens on fruit and vegetables. J. Food Prot. 2004, 67, 2839–2849. [Google Scholar] [PubMed]
- Mohamed, Z.A.; Shehri, A.M.A. Microcystins in groundwater wells and their accumulation in vegetable plants irrigated with contaminated waters in Saudi Arabia. J. Hazard. Mater. 2009, 172, 310–315. [Google Scholar] [CrossRef] [PubMed]
- El-Din, M.N.A.; Madany, I.M.; Al-Tayaran, A.; Al-Jubair, A.H.; Gomaa, A. Trends in water quality of some wells in Saudi Arabia, 1984–1989. Sci. Total Environ. 1994, 143, 173–181. [Google Scholar] [CrossRef]
- AlOtaibi, E.L. Bacteriological assessment of urban water sources in Khamis Mushait governorate, southwestern Saudi Arabia. Int. J. Health Geogr. 2009, 8. [Google Scholar] [CrossRef] [PubMed]
- Al-Salamah, I.; Nassar, I.N. Trends in drinking water quality for some wells in Qassim, Saudi Arabia, 1997–2009. J. Appl. Sci. 2009, 9, 3555–3561. [Google Scholar] [CrossRef]
- Al-Salamah, I.S.; Ghazaw, Y.M.; Ghumman, A.R. Groundwater modeling of Saq Buraydah Al Qassim for better water management strategies. Environ. Monit. Assess. 2011, 173, 851–860. [Google Scholar] [CrossRef] [PubMed]
- KAUST Industry Collaboration Program (KICP). Promoting Wastewater Reclamation and Reuse in the Kingdom of Saudi Arabia: Technology Trends, Innovation Needs, and Business Opportunities; KAUST Industry Collaboration Program (KICP): Thuwal, Saudi Arabia, 2010. [Google Scholar]
- Missimer, T.M.; Drewes, J.E.; Amy, G.; Maliva, R.G.; Keller, S. Restoration of wadi aquifers by artificial recharge with treated waste water. Groundwater 2012, 50, 514–527. [Google Scholar] [CrossRef] [PubMed]
- APHA; AWWA; WEF. Standard Method for the Examination of Water and Wastewater; American Water Works Association: Denver, CO, USA, 2012; pp. 9–69. [Google Scholar]
- Ansari, M.I.; Harb, M.; Jones, B.; Hong, P.Y. Molecular-based approaches to characterize coastal microbial community and their potential relation to the trophic state of Red Sea. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Al-Jassim, N.; Ansari, M.I.; Harb, M.; Hong, P.Y. Removal of bacterial contaminants and antibiotic resistance genes by conventional wastewater treatment processes in Saudi Arabia: Is the treated wastewater safe to reuse for agricultural irrigation? Water Res. 2015, 73, 277–290. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Kelly, W.R.; Panno, S.V.; Liu, W.T. Tracing fecal pollution sources in karst groundwater by Bacteroidales genetic biomarkers, bacterial indicators, and environmental variables. Sci. Total Environ. 2014, 490, 1082–1090. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Santo Domingo, J.; Shanks, O.C. Identification of chicken-specific fecal microbial sequences using a metagenomic approach. Water Res. 2007, 41, 3561–3574. [Google Scholar] [CrossRef] [PubMed]
- Cole, J.R.; Wang, Q.; Cardenas, E.; Fish, J.; Chai, B.; Farris, R.J.; Kulam-Syed-Mohideen, A.S.; McGarrell, D.M.; Marsh, T.; Garrity, G.M.; et al. The Ribosomal Database Project: Improved alignments and new tools for rrna analysis. Nucleic. Acids. Res. 2009, 37, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Clarke, K.; Gorley, R. Primer Version 7: User Manual/Tutorial; PRIMER-E: Plymouth, UK, 2015. [Google Scholar]
- Clarke, K.R.; Somerfield, P.J.; Gorley, R.N. Testing of null hypotheses in exploratory community analyses: Similarity profiles and biota-environment linkage. J. Exp. Mar. Biol. Ecol. 2008, 366, 56–69. [Google Scholar] [CrossRef]
- McKenna, M. Antibiotic resistance: The last resort. Nature 2013, 499, 394–396. [Google Scholar] [CrossRef] [PubMed]
- Zowawi, H.M.; Balkhy, H.H.; Walsh, T.R.; Paterson, D.L. Beta-lactamase production in key gram-negative pathogen isolates from the Arabian peninsula. Clin. Microbiol. Rev. 2013, 26, 361–380. [Google Scholar] [CrossRef] [PubMed]
- Gerba, C.; Choi, C. Role of Irrigation Water in Crop Contamination by Viruses. In Viruses in Foods; Goyal, S., Ed.; Springer US: New York, NY, USA, 2006; pp. 257–263. [Google Scholar]
- USEPA. Exposure Factors Handbook 2011; U.S. Environmental Protection Agency: Washington, DC, USA, 2011.
- Dupont, H.; Montravers, P.; Mohler, J.; Carbon, C. Disparate findings on the role of virulence factors of Enterococcus faecalis in mouse and rat models of peritonitis. Infect. Immunol. 1998, 66, 2570–2575. [Google Scholar]
- Schook, L.B.; Carrick, L.; Berk, R.S. Murine gastrointestinal-tract as a portal of entry in experimental Pseudomonas aeruginosa infections. Infect. Immunol. 1976, 14, 564–570. [Google Scholar]
- Smeets, P.W.M.H.; Medema, G.J.; van Dijk, J.C. The Dutch secret: How to provide safe drinking water without chlorine in the Netherlands. Drink Water Eng. Sci. 2009, 2, 1–14. [Google Scholar] [CrossRef]
- Chowdhury, S.; Al-Zahrani, M. Characterizing water resources and trends of sector wise water consumptions in Saudi Arabia. Eng. Sci. 2015, 27, 68–82. [Google Scholar] [CrossRef]
- FAO. Irrigation in the Middle East Region in Figures; Food and Agriculture Organization of the United Nations: Rome, Italy, 2009. [Google Scholar]
- Al-Jasser, A.O. Saudi wastewater reuse standards for agricultural irrigation: Riyadh treatment plants effluent compliance. Eng. Sci. 2011, 23, 1–8. [Google Scholar] [CrossRef]
- O’Sullivan, E. Saudi Arabia Presses Ahead with a Wastewater Revolution; MEED: London, UK, 2010. [Google Scholar]
- Al-Sefry, S.A.; Şen, Z. Groundwater rise problem and risk evaluation in major cities of arid lands—Jedddah case in Kingdom of Saudi Arabia. Water Resour. Manag. 2006, 20, 91–108. [Google Scholar] [CrossRef]
- Hong, P.Y.; Yannarell, A.C.; Dai, Q.; Ekizoglu, M.; Mackie, R.I. Monitoring the perturbation of soil and groundwater microbial communities due to pig production activities. Appl. Environ. Microbiol. 2013, 79, 2620–2629. [Google Scholar] [CrossRef] [PubMed]
- Francis, C.A.; Roberts, K.J.; Beman, J.M.; Santoro, A.E.; Oakley, B.B. Ubiquity and diversity of ammonia-oxidizing archaea in water columns and sediments of the ocean. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 14683–14688. [Google Scholar] [CrossRef] [PubMed]
- Leininger, S.; Urich, T.; Schloter, M.; Schwark, L.; Qi, J.; Nicol, G.W.; Prosser, J.I.; Schuster, S.C.; Schleper, C. Archaea predominate among ammonia-oxidizing prokaryotes in soils. Nature 2006, 442, 806–809. [Google Scholar] [CrossRef] [PubMed]
- Van der Wielen, P.W.J.J.; Voost, S.; van der Kooij, D. Ammonia-oxidizing bacteria and archaea in groundwater treatment and drinking water distribution systems. Appl. Environ. Microbiol. 2009, 75, 4687–4695. [Google Scholar] [CrossRef] [PubMed]
- Johnston, E.L.; Roberts, D.A. Contaminants reduce the richness and evenness of marine communities: A review and meta-analysis. Environ. Pollut. 2009, 157, 1745–1752. [Google Scholar] [CrossRef] [PubMed]
- Hooper, D.U.; Chapin, F.S.; Ewel, J.J.; Hector, A.; Inchausti, P.; Lavorel, S.; Lawton, J.H.; Lodge, D.M.; Loreau, M.; Naeem, S.; et al. Effects of biodiversity on ecosystem functioning: A consensus of current knowledge. Ecol. Monogr. 2005, 75, 3–35. [Google Scholar] [CrossRef]
- Hong, P.Y.; Wu, J.H.; Liu, W.T. Relative abundance of Bacteroides spp. In stools and wastewaters as determined by hierarchical oligonucleotide primer extension. Appl. Environ. Microbiol. 2008, 74, 2882–2893. [Google Scholar] [CrossRef] [PubMed]
- Kreader, C.A. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl. Environ. Microbiol. 1995, 61, 1171–1179. [Google Scholar] [PubMed]
- Hong, P.Y.; Wu, J.H.; Liu, W.T. A high-throughput and quantitative hierarchical oligonucleotide primer extension (hope)-based approach to identify sources of faecal contamination in water bodies. Environ. Microbiol. 2009, 11, 1672–1681. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Doyle, M.P. Survival of enterohemorrhagic Escherichia coli O157:H7 in water. J. Food. Prot. 1998, 61, 662–667. [Google Scholar] [PubMed]
- Dong, S.; Hong, P.Y.; Nguyen, T.H. Persistence of Bacteroides ovatus under simulated sunlight irradiation. BMC. microbiol. 2014, 14. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aly, M.; Balkhy, H.H. The prevalence of antimicrobial resistance in clinical isolates from gulf corporation council countries. Antimicrob. Resist. Infect. Control 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Memish, Z.A.; Shibl, A.M.; Kambal, A.M.; Ohaly, Y.A.; Ishaq, A.; Livermore, D.M. Antimicrobial resistance among non-fermenting gram-negative bacteria in Saudi Arabia. J. Antimicrob. Chemother. 2012, 67, 1701–1705. [Google Scholar] [CrossRef] [PubMed]
- Peleg, A.Y.; Seifert, H.; Paterson, D.L. Acinetobacter baumannii: Emergence of a successful pathogen. Clin. Microbiol. Rev. 2008, 21, 538–582. [Google Scholar] [CrossRef] [PubMed]
- Stoddard, S.F.; Smith, B.J.; Hein, R.; Roller, B.R.; Schmidt, T.M. Rrndb: Improved tools for interpreting rRNA gene abundance in bacteria and archaea and a new foundation for future development. Nucleic. Acids. Res. 2015, 43, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Farrelly, V.; Rainey, F.A.; Stackebrandt, E. Effect of genome size and rrn gene copy number on PCR amplification of 16s rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 1995, 61, 2798–2801. [Google Scholar] [PubMed]
- Crosby, L.D.; Criddle, C.S. Understanding bias in microbial community analysis techniques due to rrn operon copy number heterogeneity. BioTechniques. 2003, 34, 790–794. [Google Scholar] [PubMed]
- Toranzo, A.E.; Magarinos, B.; Romalde, J.L. A review of the main bacterial fish diseases in mariculture systems. Aquaculture 2005, 246, 37–61. [Google Scholar] [CrossRef]
- Hassan, S.A.; Altalhi, A.D.; Gherbawy, Y.A.; El-Deeb, B.A. Bacterial load of fresh vegetables and their resistance to the currently used antibiotics in Saudi Arabia. Foodborne Pathoge. Dis. 2011, 8, 1011–1018. [Google Scholar] [CrossRef] [PubMed]
- Marti, R.; Scott, A.; Tien, Y.C.; Murray, R.; Sabourin, L.; Zhang, Y.; Topp, E. Impact of manure fertilization on the abundance of antibiotic-resistant bacteria and frequency of detection of antibiotic resistance genes in soil and on vegetables at harvest. Appl. Environ. Microb. 2013, 79, 5701–5709. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alsalah, D.; Al-Jassim, N.; Timraz, K.; Hong, P.-Y. Assessing the Groundwater Quality at a Saudi Arabian Agricultural Site and the Occurrence of Opportunistic Pathogens on Irrigated Food Produce. Int. J. Environ. Res. Public Health 2015, 12, 12391-12411. https://doi.org/10.3390/ijerph121012391
Alsalah D, Al-Jassim N, Timraz K, Hong P-Y. Assessing the Groundwater Quality at a Saudi Arabian Agricultural Site and the Occurrence of Opportunistic Pathogens on Irrigated Food Produce. International Journal of Environmental Research and Public Health. 2015; 12(10):12391-12411. https://doi.org/10.3390/ijerph121012391
Chicago/Turabian StyleAlsalah, Dhafer, Nada Al-Jassim, Kenda Timraz, and Pei-Ying Hong. 2015. "Assessing the Groundwater Quality at a Saudi Arabian Agricultural Site and the Occurrence of Opportunistic Pathogens on Irrigated Food Produce" International Journal of Environmental Research and Public Health 12, no. 10: 12391-12411. https://doi.org/10.3390/ijerph121012391
APA StyleAlsalah, D., Al-Jassim, N., Timraz, K., & Hong, P.-Y. (2015). Assessing the Groundwater Quality at a Saudi Arabian Agricultural Site and the Occurrence of Opportunistic Pathogens on Irrigated Food Produce. International Journal of Environmental Research and Public Health, 12(10), 12391-12411. https://doi.org/10.3390/ijerph121012391






