Effects of Gold Nanorods on Imprinted Genes Expression in TM-4 Sertoli Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Regents
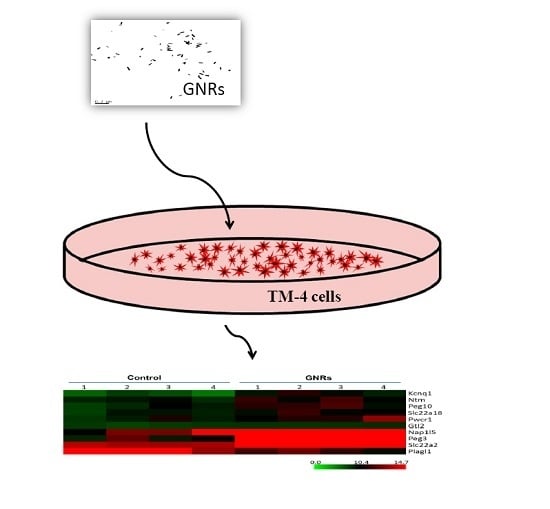
2.2. Cell Culture and GNRs Treatment
2.3. Characteristics of GNRs
2.4. Total RNA Isolation
2.5. Real-Time PCR
2.6. Expression Level of Imprinted Genes
2.7. Statistical Analysis
3. Results
3.1 Expression of Imprinted Genes in Normal and GNRs Treated Group
3.2. Different Expression of Imprinted Genes between Expression Allele
3.3. Enriched Biological Trends within the Imprinted Gene Set
4. Discussion
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| GNRs | Gold Nanorods |
| ICR | Imprinting Control Regions |
| MXC | Methoxychlor |
References
- Akiyama, Y.; Mori, T.; Katayama, Y.; Niidome, T. Conversion of rod-shaped gold nanoparticles to spherical forms and their effect on biodistribution in tumor-bearing mice. Nanoscale Res. Lett. 2012, 7, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Heidari, Z.; Salouti, M.; Sariri, R. Breast cancer photothermal therapy based on gold nanorods targeted by covalently-coupled bombesin peptide. Nanotechnology 2015, 26, 195101. [Google Scholar] [CrossRef] [PubMed]
- Shang, T.; Wang, C.D.; Ren, L.; Tian, X.H.; Li, D.H.; Ke, X.B.; Chen, M.; Yang, A.Q. Synthesis and characterization of nir-responsive aurod@pnipaam-pegma nanogels as vehicles for delivery of photodynamic therapy agents. Nanoscale Res. Lett. 2013, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Xia, K.; Bai, Y.Y.; Tang, Y.; Deng, Y.; Chen, J.; Qian, W.; Shen, H.; Zhang, Z.; Ju, S.; et al. Synthesis of gold nanorods and their functionalization with bovine serum albumin for optical hyperthermia. J. Biomed. Nanotechnol. 2014, 10, 1440–1449. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.K.; Emoto, K.; Su, L.J.; Yang, X.; Flaig, T.W.; Park, W. Functionalized gold nanorods for thermal ablation treatment of bladder cancer. J. Biomed. Nanotechnol. 2014, 10, 1267–1276. [Google Scholar] [CrossRef] [PubMed]
- Kondath, S.; Raghavan, B.S.; Anantanarayanan, R.; Rajaram, R. Synthesis and characterisation of morin reduced gold nanoparticles and its cytotoxicity in mcf-7 cells. Chem. Biol. Interact. 2014, 224, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.H.; Kim, S.J.; Kim, Y.H.; Noh, J.R.; Gang, G.T.; Chung, B.H.; Song, N.W.; Lee, C.H. Susceptibility to gold nanoparticle-induced hepatotoxicity is enhanced in a mouse model of nonalcoholic steatohepatitis. Toxicology 2012, 294, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Meng, J.; Ji, Y.; Liu, J.; Cheng, X.; Guo, H.; Zhang, W.; Wu, X.; Xu, H. Using gold nanorods core/silver shell nanostructures as model material to probe biodistribution and toxic effects of silver nanoparticles in mice. Nanotoxicology 2014, 8, 686–696. [Google Scholar] [CrossRef] [PubMed]
- Malugin, A.; Ghandehari, H. Cellular uptake and toxicity of gold nanoparticles in prostate cancer cells: A comparative study of rods and spheres. J. Appl. Toxicol. 2010, 30, 212–217. [Google Scholar] [PubMed]
- Mahmood, M.; Casciano, D.A.; Mocan, T.; Iancu, C.; Xu, Y.; Mocan, L.; Iancu, D.T.; Dervishi, E.; Li, Z.; Abdalmuhsen, M.; et al. Cytotoxicity and biological effects of functional nanomaterials delivered to various cell lines. J. Appl. Toxicol. 2010, 30, 74–83. [Google Scholar] [CrossRef] [PubMed]
- Wiwanitkit, V.; Sereemaspun, A.; Rojanathanes, R. Effect of gold nanoparticles on spermatozoa: The first world report. Fertil. Steril. 2009, 91, e7–e8. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Chen, M.; Ji, X.; Mao, Z.; Zhang, X.; Wang, X.; Xia, Y. Metabolomic profiles delineate the potential role of glycine in gold nanorod-induced disruption of mitochondria and blood-testis barrier factors in tm-4 cells. Nanoscale 2014, 6, 8265–8273. [Google Scholar] [CrossRef] [PubMed]
- Lambertini, L. Genomic imprinting: Sensing the environment and driving the fetal growth. Curr. Opin. Pediatr. 2014, 26, 237–242. [Google Scholar] [CrossRef] [PubMed]
- Barlow, D.P. Genomic imprinting: A mammalian epigenetic discovery model. Annu. Rev. Genet. 2011, 45, 379–403. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, W.A.; Mann, M.R. Epigenetic regulation of genomic imprinting from germ line to preimplantation. Mol. Reprod. Dev. 2014, 81, 126–140. [Google Scholar] [CrossRef] [PubMed]
- Shamanski, F.L.; Kimura, Y.; Lavoir, M.C.; Pedersen, R.A.; Yanagimachi, R. Status of genomic imprinting in mouse spermatids. Hum. Reprod. 1999, 14, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Boissonnas, C.C.; Abdalaoui, H.E.; Haelewyn, V.; Fauque, P.; Dupont, J.M.; Gut, I.; Vaiman, D.; Jouannet, P.; Tost, J.; Jammes, H. Specific epigenetic alterations of igf2-h19 locus in spermatozoa from infertile men. Eur. J. Hum. Genet. 2010, 18, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Uyar, A.; Seli, E. The impact of assisted reproductive technologies on genomic imprinting and imprinting disorders. Curr. Opin. Obstet. Gynecol. 2014, 26, 210–221. [Google Scholar] [CrossRef] [PubMed]
- Diplas, A.I.; Lambertini, L.; Lee, M.J.; Sperling, R.; Lee, Y.L.; Wetmur, J.; Chen, J. Differential expression of imprinted genes in normal and iugr human placentas. Epigenetics 2009, 4, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Smeester, L.; Yosim, A.E.; Nye, M.D.; Hoyo, C.; Murphy, S.K.; Fry, R.C. Imprinted genes and the environment: Links to the toxic metals arsenic, cadmium, lead and mercury. Genes 2014, 5, 477–496. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.J.; Carvalho, F.; Sousa, M.; Barros, A. Genomic imprinting in disruptive spermatogenesis. Lancet 2004, 363, 1700–1702. [Google Scholar] [CrossRef]
- Cui, H. Loss of imprinting of igf2 as an epigenetic marker for the risk of human cancer. Dis. Markers 2007, 23, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Marques, C.J.; Costa, P.; Vaz, B.; Carvalho, F.; Fernandes, S.; Barros, A.; Sousa, M. Abnormal methylation of imprinted genes in human sperm is associated with oligozoospermia. Mol. Hum. Reprod. 2008, 14, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Kang, E.-R.; Iqbal, K.; Tran, D.A.; Rivas, G.E.; Singh, P.; Pfeifer, G.P.; Szabó, P.E. Effects of endocrine disruptors on imprinted gene expression in the mouse embryo. Epigenetics 2011, 6, 937–950. [Google Scholar] [CrossRef] [PubMed]
- LaRocca, J.; Binder, A.M.; McElrath, T.F.; Michels, K.B. The impact of first trimester phthalate and phenol exposure on igf2/h19 genomic imprinting and birth outcomes. Environ. Res. 2014, 133, 396–406. [Google Scholar] [CrossRef] [PubMed]
- Badcock, C.; Crespi, B. Imbalanced genomic imprinting in brain development: An evolutionary basis for the aetiology of autism. J. Evol. Biol. 2006, 19, 1007–1032. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.B.; Bartolomei, M.S. Regulation of imprinting in clusters: Noncoding RNAs versus insulators. Adv. Genet. 2008, 61, 207–223. [Google Scholar] [PubMed]
- De Crescenzo, A.; Sparago, A.; Cerrato, F.; Palumbo, O.; Carella, M.; Miceli, M.; Bronshtein, M.; Riccio, A.; Yaron, Y. Paternal deletion of the 11p15.5 centromeric-imprinting control region is associated with alteration of imprinted gene expression and recurrent severe intrauterine growth restriction. J. Med. Genet. 2013, 50, 99–103. [Google Scholar] [CrossRef] [PubMed]
- De la Casa Esperon, E.; Cordier, G.; Engel, N. A genomic reservoir for tnfrsf genes is developmentally regulated and imprinted in the mouse. Epigenetics 2012, 7, 626–634. [Google Scholar] [CrossRef] [PubMed]
- Erickson, R.P. Cattanach’s translocation [is(7:X)ct] corrects male sterility due to homozygosity for chromosome 7 deletions. Genet. Res. 1984, 43, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Gianotten, J.; van der Veen, F.; Alders, M.; Leschot, N.J.; Tanck, M.W.; Land, J.A.; Kremer, J.A.; Hoefsloot, L.H.; Mannens, M.M.; Lombardi, M.P.; et al. Chromosomal region 11p15 is associated with male factor subfertility. Mol. Hum. Reprod. 2003, 9, 587–592. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Liu, Y.; Sun, M.; Niu, M.; Yuan, Q.; Hai, Y.; Guo, Y.; Chen, Z.; Hou, J.; He, Z. Micrornas and DNA methylation as epigenetic regulators of mitosis, meiosis and spermiogenesis. Reproduction 2015, 150, R25–R34. [Google Scholar] [CrossRef] [PubMed]
- Stoccoro, A.; Karlsson, H.L.; Coppede, F.; Migliore, L. Epigenetic effects of nano-sized materials. Toxicology 2013, 313, 3–14. [Google Scholar] [CrossRef] [PubMed]
- Tsevi, I.; Vicente, R.; Grande, M.; Lopez-Iglesias, C.; Figueras, A.; Capella, G.; Condom, E.; Felipe, A. Kcnq1/kcne1 channels during germ-cell differentiation in the rat: Expression associated with testis pathologies. J. Cell. Physiol. 2005, 202, 400–410. [Google Scholar] [CrossRef] [PubMed]
- Rajender, S.; Avery, K.; Agarwal, A. Epigenetics, spermatogenesis and male infertility. Mutat. Res. 2011, 727, 62–71. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sun, M.; Zhao, G.; Liu, J.; Gao, W.; Si, S.; Meng, T. Elevated expression of peg10 in human placentas from preeclamptic pregnancies. Acta Histochem. 2012, 114, 589–593. [Google Scholar] [CrossRef] [PubMed]
- Doria, S.; Sousa, M.; Fernandes, S.; Ramalho, C.; Brandao, O.; Matias, A.; Barros, A.; Carvalho, F. Gene expression pattern of igf2, phlda2, peg10 and cdkn1c imprinted genes in spontaneous miscarriages or fetal deaths. Epigenetics 2010, 5, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Onteru, S.K.; Fan, B.; Nikkila, M.T.; Garrick, D.J.; Stalder, K.J.; Rothschild, M.F. Whole-genome association analyses for lifetime reproductive traits in the pig. J. Anim. Sci. 2011, 89, 988–995. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.X.; Sun, B.F.; Jiao, J.; Chong, Z.C.; Chen, Y.S.; Wang, X.L.; Zhao, Y.; Zhou, Y.M.; Li, D. Genome-wide 5-hydroxymethylcytosine modification pattern is a novel epigenetic feature of globozoospermia. Oncotarget 2015, 6, 6535–6543. [Google Scholar] [CrossRef] [PubMed]
- El Hajj, N.; Zechner, U.; Schneider, E.; Tresch, A.; Gromoll, J.; Hahn, T.; Schorsch, M.; Haaf, T. Methylation status of imprinted genes and repetitive elements in sperm DNA from infertile males. Sex. Dev. 2011, 5, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Stouder, C.; Paoloni-Giacobino, A. Specific transgenerational imprinting effects of the endocrine disruptor methoxychlor on male gametes. Reproduction 2011, 141, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Frey, W.D.; He, H.; Kim, H.; Ekram, M.B.; Bakshi, A.; Faisal, M.; Perera, B.P.; Ye, A.; Teruyama, R. Peg3 mutational effects on reproduction and placenta-specific gene families. PLoS ONE 2013, 8. [Google Scholar] [CrossRef] [PubMed]




| Gene | Expression | Gene | Expression | Gene | Expression | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mouse | Human | 0 nM | 10 nM | Mouse | Human | 0 nM | 10 nM | Mouse | Human | 0 nM | 10 nM |
| Ano1 | ANO1 | Yes | No | Kcnk9 | KCNK9 | Yes | Yes | Phlda2 | PHLDA2 | Yes | Yes |
| Cdkn1c | CDKN1C | No | No | Kcnq1 | KCNQ1 | Yes | Yes | Plagl1 | PLAGL1 | No | Yes |
| Ddc | DDC | Yes | Yes | Klf14 | KLF14 | Yes | Yes | Ppp1r9a | PPP1R9A | No | No |
| Dlk1 | DLK1 | Yes | Yes | Magel2 | MAGEL2 | No | No | Pwcr1 | PWCR1 | Yes | Yes |
| Gnas | GNAS | Yes | Yes | Magi2 | MAGI2 | Yes | Yes | Rian | MEG8 | Yes | Yes |
| Gnas-as | GNAS-AS | Yes | Yes | Mest | MEST | No | No | Sgce | SGCE | No | No |
| Gpr1 | GPR1 | Yes | Yes | Mir296 | MIR296 | Yes | Yes | Slc22a18 | SLC22A18 | Yes | Yes |
| Grb10 | GRB10 | Yes | Yes | Mir298 | MIR298 | Yes | Yes | Slc22a2 | SLC22A2* | Yes | No |
| Gtl2 | MEG3 | Yes | Yes | Mkrn3 | MKRN3 | Yes | Yes | Slc22a3 | SLC22A3* | No | No |
| H19 | H19 | Yes | Yes | Nap1l5 | NAP1L5 | Yes | No | Snrpn | SNRPN | Yes | Yes |
| Hymai | HYMAI | No | No | Ndn | NDN | No | No | Snurf | SNURF | No | No |
| Igf2 | IGF2 | No | No | Nnat | NNAT | Yes | Yes | Tfpi2 | TFPI2 | Yes | Yes |
| Igf2as | IGF2AS | Yes | Yes | Ntm | NTM | Yes | Yes | Ube3a | UBE3A | Yes | Yes |
| Igf2r | IGF2R | Yes | Yes | Peg10 | PEG10 | Yes | Yes | Zim2 | ZIM2 | No | No |
| Inpp5f V2 | INPP5F V2 | Yes | Yes | Peg3 | PEG3 | Yes | No | ||||
| Gene | p Value | Location | Full Name | Gene Type | Expression Allele | |
|---|---|---|---|---|---|---|
| Mouse | Human | |||||
| Kcnq1 | 0.003 | 7 69.3 cM | 11p15.5 | potassium voltage-gated channel, subfamily Q, member 1 | PC | M |
| Ntm | 0.026 | 9 AS | 11q25 | neurotrimin | PC | M |
| Peg10 | 0.030 | 6 0.5 cM | 7q21 | paternally expressed 10 | PC | P |
| Slc22a18 | 0.042 | 7 69.5 cM | 11p15.5 | solute carrier family 22 (organic cation transporter), member 18 | PC | M |
| Pwcr1 | 0.046 | 7 29.0 cM AS | 15q11.2 | Prader-Willi syndrome chromosome region 1 | NC | P |
| Gtl2 | 0.048 | 12 54.0 cM | 14q32 | gene trap locus 2 | NC | M |
| Nap1l5 | 0.005 | 6 C1 AS | 4q22.1 AS | nucleosome assembly protein 1-like 5 | PC | P |
| Peg3 | <0.001 | 7 6.5 cM AS | 19q13.4 AS | paternally expressed 3 | PC | P |
| Slc22a2 | 0.013 | 17 7.32 cM | 6q26 AS | solute carrier family 22 (organic cation transporter), member 2 | PC | M |
| Plagl1 | 0.019 | 10 15.0 cM | 6q24-q25 AS | pleiomorphic adenoma gene-like 1 | PC | P |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, B.; Gu, H.; Xu, B.; Tang, Q.; Wu, W.; Ji, X.; Xia, Y.; Hu, L.; Chen, D.; Wang, X. Effects of Gold Nanorods on Imprinted Genes Expression in TM-4 Sertoli Cells. Int. J. Environ. Res. Public Health 2016, 13, 271. https://doi.org/10.3390/ijerph13030271
Yuan B, Gu H, Xu B, Tang Q, Wu W, Ji X, Xia Y, Hu L, Chen D, Wang X. Effects of Gold Nanorods on Imprinted Genes Expression in TM-4 Sertoli Cells. International Journal of Environmental Research and Public Health. 2016; 13(3):271. https://doi.org/10.3390/ijerph13030271
Chicago/Turabian StyleYuan, Beilei, Hao Gu, Bo Xu, Qiuqin Tang, Wei Wu, Xiaoli Ji, Yankai Xia, Lingqing Hu, Daozhen Chen, and Xinru Wang. 2016. "Effects of Gold Nanorods on Imprinted Genes Expression in TM-4 Sertoli Cells" International Journal of Environmental Research and Public Health 13, no. 3: 271. https://doi.org/10.3390/ijerph13030271