Microbial Insight into a Pilot-Scale Enhanced Two-Stage High-Solid Anaerobic Digestion System Treating Waste Activated Sludge
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Set-Up
2.2. Inoculum and Feed
2.3. Analytical Methods
2.4. Microbial Community Analysis
3. Results and Discussion
3.1. Performance of Pilot HSAD System
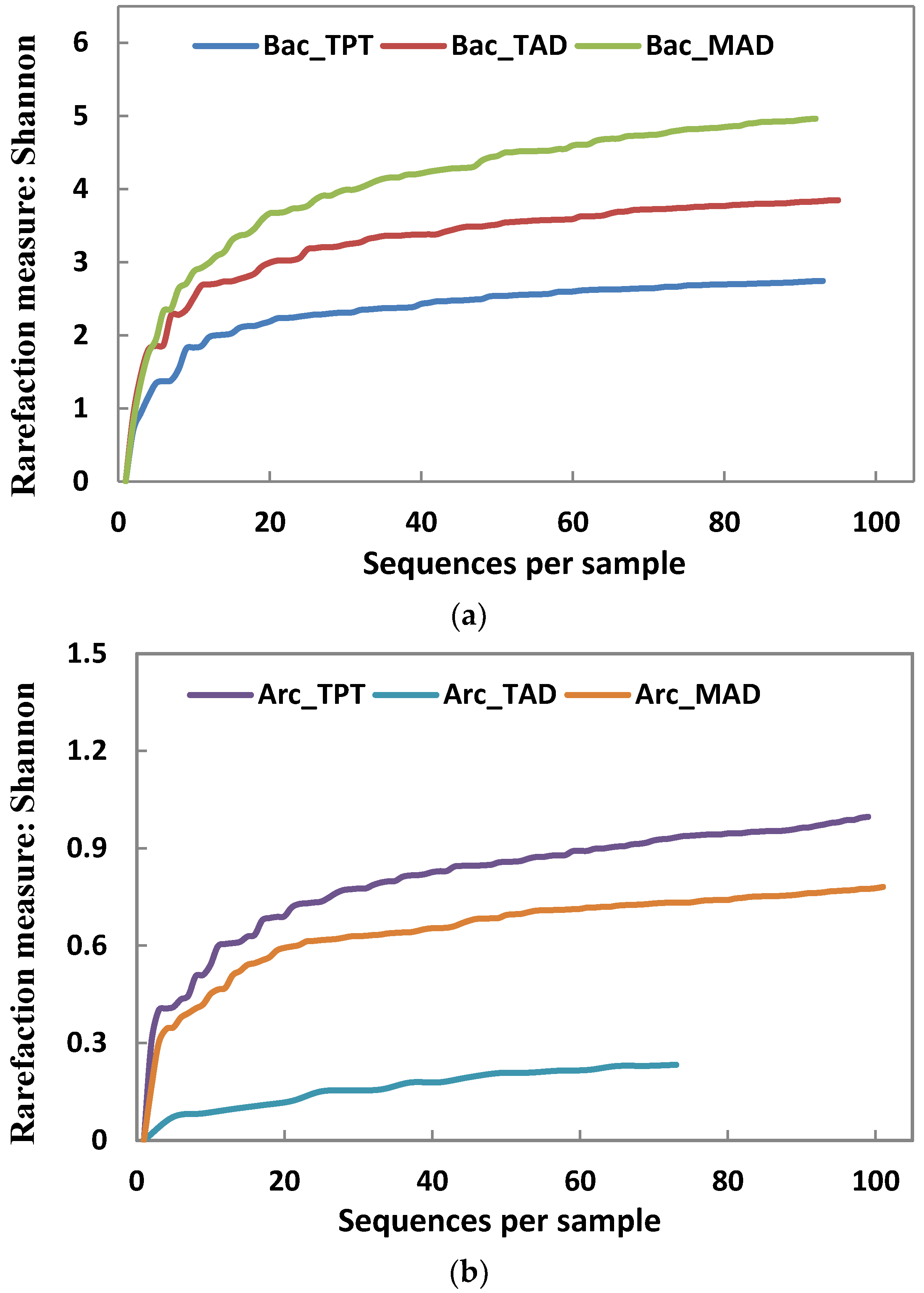
3.2. Microbial Community Structures
3.3. Composition of Microbial Communities
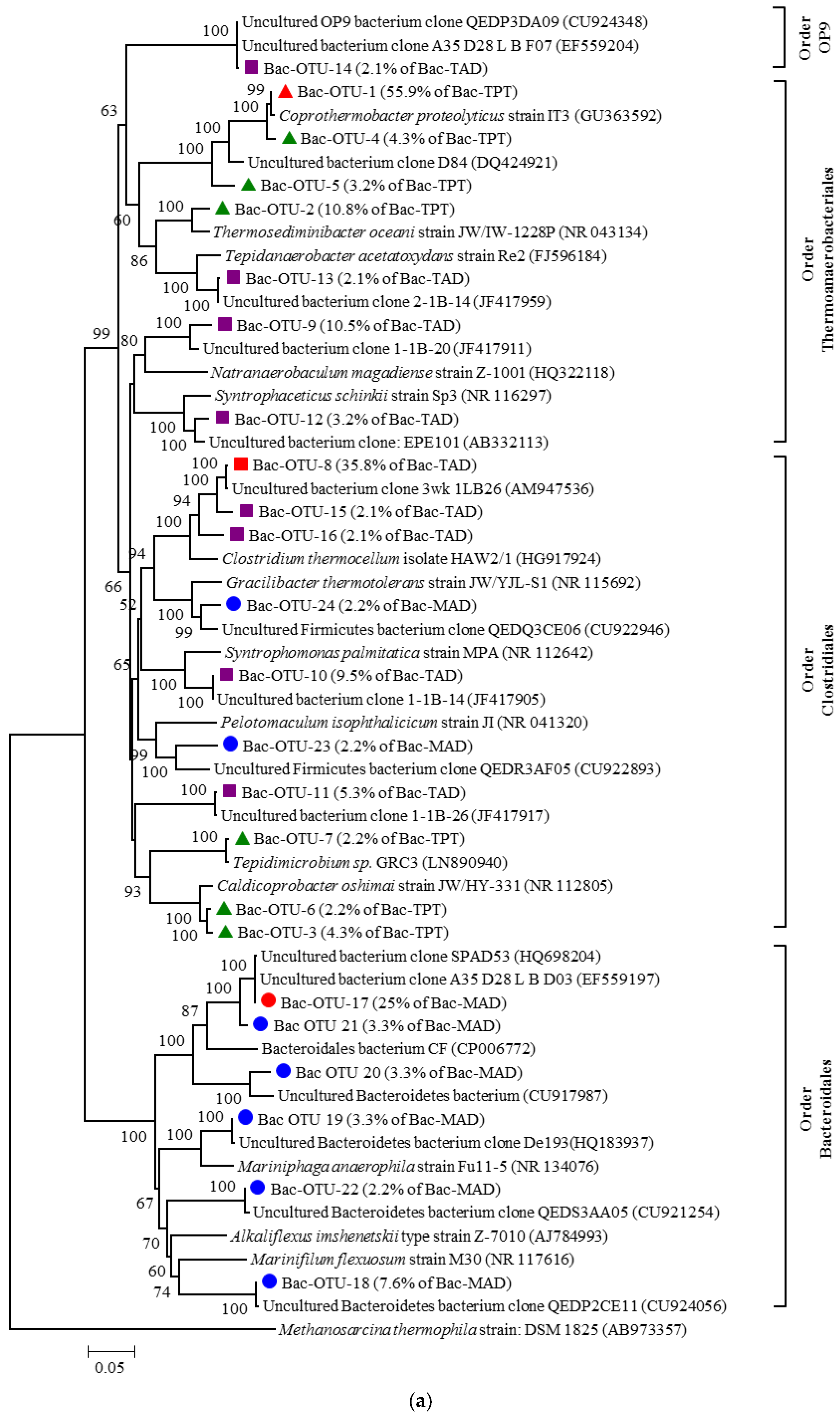
3.3.1. Composition of Bacterial Communities
Composition of Bac-TPT Library
Composition of Bac-TAD Library
Composition of Bac-MAD Library
3.3.2. Composition of Archaeal Communities
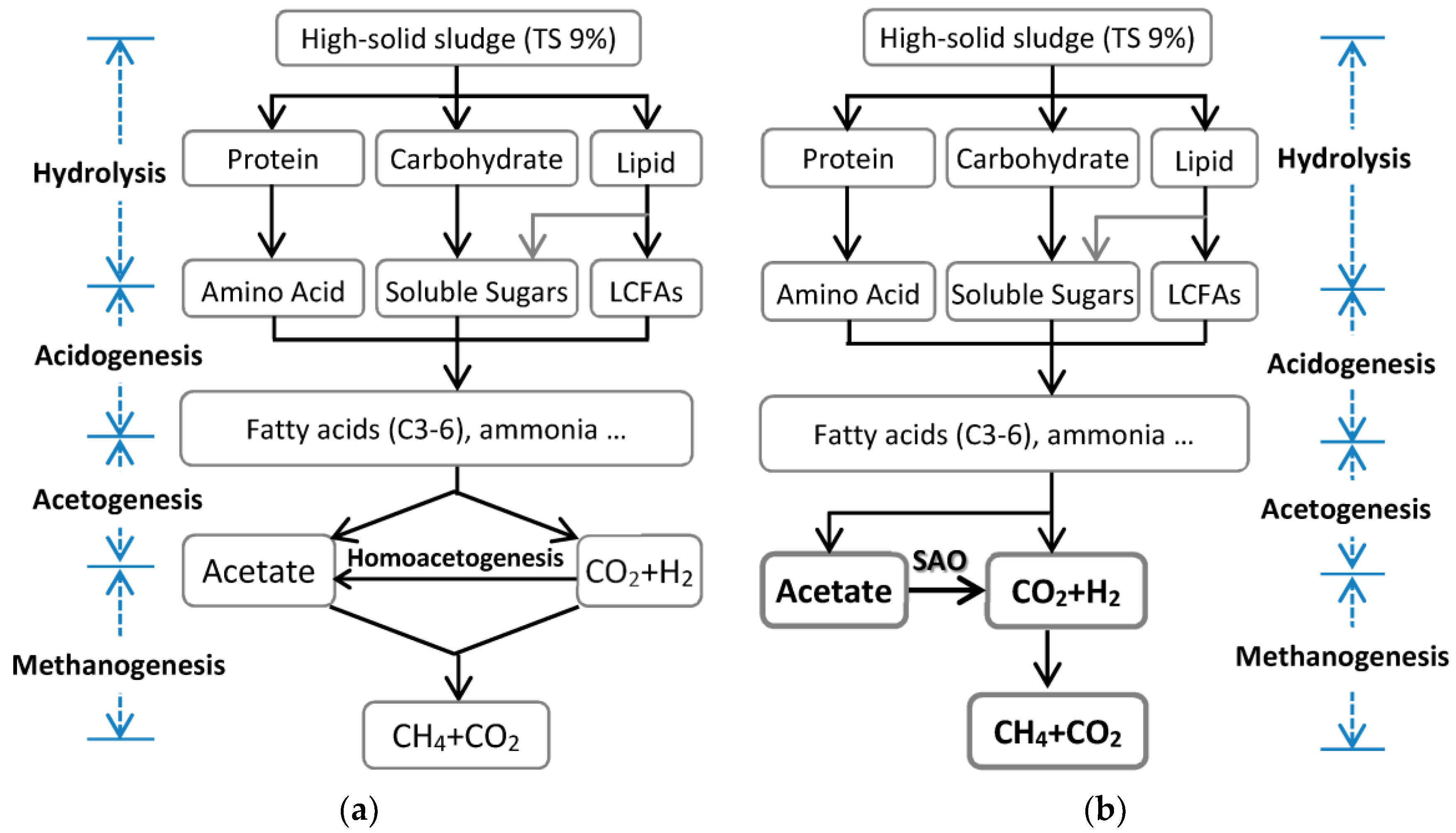
3.4. Possible Conversion Route of Organics of Sludge
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Duan, N.; Dong, B.; Wu, B.; Dai, X. High-solid anaerobic digestion of sewage sludge under mesophilic conditions: Feasibility study. Bioresour. Technol. 2012, 104, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Baudez, J.C.; Gupta, R.K.; Eshtiaghi, N.; Slatter, P. The viscoelastic behaviour of raw and anaerobic digested sludge: Strong similarities with soft-glassy materials. Water Res. 2013, 47, 173–180. [Google Scholar] [CrossRef] [PubMed]
- US Environmental Protection Agency. Land Application of Sewage Sludge. A Guide for Land Appliers on the Requirements of the Federal Standards for the Use or Disposal of Sewage; 40 CFR Part 503; US Environmental Protection Agency: Washington, DC, USA, 1994.
- Abbassi-Guendouz, A.; Brockmann, D.; Trably, E.; Dumas, C.; Delgenes, J.P.; Steyer, J.P.; Escudié, R. Total solids content drives high solid anaerobic digestion via mass transfer limitation. Bioresour. Technol. 2012, 111, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Li, N.; Dong, B.; Zhao, W.; Dai, L.; Dai, X. New insights into the enhanced performance of high solid anaerobic digestion with dewatered sludge by thermal hydrolysis: Organic matter degradation and methanogenic pathways. J. Hazard. Mater. 2018, 342, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Lee, C.Y.; Chang, S.W.; Kim, D.J. Enhanced methane production and wastewater sludge stabilization of a continuous full scale thermal pretreatment and thermophilic anaerobic digestion. Bioresour. Technol. 2017, 245, 1162–1167. [Google Scholar] [CrossRef] [PubMed]
- Zinder, S.; Koch, M. Non-aceticlastic methanogenesis from acetate: Acetate oxidation by a thermophilic syntrophic coculture. Arch. Microbiol. 1984, 138, 263–272. [Google Scholar] [CrossRef]
- Fujishima, S.; Miyahara, T.; Noike, T. Effect of moisture content on anaerobic digestion of dewatered sludge: Ammonia inhibition to carbohydrate removal and methane production. Water Sci. Technol. 2000, 41, 119–127. [Google Scholar] [PubMed]
- Liu, C.; Li, H.; Zhang, Y.; Si, D.; Chen, Q. Evolution of microbial community along with increasing solid concentration during high-solids anaerobic digestion of sewage sludge. Bioresour. Technol. 2016, 216, 87–94. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.X.; Ma, X.Y.; Wang, M.M.; Chu, H.J.; Zuo, J.E.; Yang, Y.F. A comparative study of microbial community compositions in thermophilic and mesophilic sludge anaerobic digestion systems. Microbiol. China 2016, 43, 26–35. [Google Scholar]
- Xu, F.; Wang, Z.W.; Tang, L.; Li, Y. A mass diffusion-based interpretation of the effect of total solids content on solid-state anaerobic digestion of cellulosic biomass. Bioresour. Technol. 2014, 167, 178–185. [Google Scholar] [CrossRef] [PubMed]
- Di Maria, F.; Barratta, M.; Bianconi, F.; Placidi, P.; Passeri, D. Solid anaerobic digestion batch with liquid digestate recirculation and wet anaerobic digestion of organic waste: Comparison of system performances and identification of microbial guilds. Waste Manag. 2017, 59, 172–180. [Google Scholar] [CrossRef] [PubMed]
- Rivière, D.; Desvignes, V.; Pelletier, E.; Chaussonnerie, S.; Guermazi, S.; Weissenbach, J.; Li, T.; Camacho, P.; Sghir, A. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009, 3, 700–714. [Google Scholar] [CrossRef] [PubMed]
- Sundberg, C.; Al-Soud, W.A.; Larsson, M.; Alm, E.; Yekta, S.S.; Svensson, B.H.; Sørensen, S.J.; Karlsson, A. 454 pyrosequencing analyses of bacterial and archaeal richness in 21 full-scale biogas digesters. FEMS Microbiol. Ecol. 2013, 85, 612–626. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, D.; Hori, T.; Haruta, S.; Ueno, Y.; Ishii, M.; Igarashi, Y. Methanogenic pathway and community structure in a thermophilic anaerobic digestion process of organic solid waste. J. Biosci. Bioeng. 2011, 111, 41–46. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Dolfing, J.; Sherry, A.; Gray, N.D.; Head, I.M.; Schnurer, A. Quantification of syntrophic acetate-oxidizing microbial communities in biogas processes. Environ. Microbiol. Rep. 2011, 3, 500–505. [Google Scholar] [CrossRef] [PubMed]
- Gomez, E.; Martin, J.; Michel, F.C. Effects of organic loading rate on reactor performance and archaeal community structure in mesophilic anaerobic digesters treating municipal sewage sludge. Waste Manag. Res. 2011, 29, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Maspolim, Y.; Zhou, Y.; Guo, C.; Xiao, K.; Ng, W.J. Comparison of single-stage and two-phase anaerobic sludge digestion systems—Performance and microbial community dynamics. Chemosphere 2014, 140, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Jiang, Y.; Cao, Z.P.; Li, Z.H.; Hu, Y.Y.; Li, H.Z.; Zuo, J.E.; Wang, K.J. Enhanced anaerobic digestion of waste activated sludge of low organic content in a novel digester. Water Sci. Technol. 2015, 72, 966–973. [Google Scholar] [CrossRef] [PubMed]
- Ministry of Environmental Protection, China. Standard Methods for the Examination of Water and Wastewater; China Environmental Science Press: Beijing, China, 2002.
- Hansen, K.H.; Angelidaki, I.; Ahring, B.K. Anaerobic Digestion of Swine Manure: Inhibition by Ammonia. Water Res. 1998, 32, 5–12. [Google Scholar] [CrossRef]
- Wang, C.; Zuo, J.; Chen, X.; Xing, W.; Xing, L.; Li, P.; Lu, X.; Li, C. Microbial community structures in an integrated two-phase anaerobic bioreactor fed by fruit vegetable wastes and wheat straw. J. Environ. Sci. 2014, 26, 2484–2492. [Google Scholar] [CrossRef] [PubMed]
- Marchesi, J.R.; Sato, T.; Weightman, A.J.; Martin, T.A.; Fry, J.C.; Hiom, S.J.; Dymock, D.; Wade, W.G. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl. Environ. Microbiol. 1998, 64, 795–799. [Google Scholar] [PubMed]
- Grosskopf, R.; Janssen, P.H.; Liesack, W. Diversity and structure of the methanogenic community in anoxic rice paddy soil microcosms as examined by cultivation and direct 16S rRNA gene sequence retrieval. Appl. Environ. Microbiol. 1998, 64, 960–969. [Google Scholar] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular Evolutionary Genetics Analysis Using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Ge, H.; Jensen, P.D.; Batstone, D.J. Temperature phased anaerobic digestion increases apparent hydrolysis rate for waste activated sludge. Water Res. 2011, 45, 1597–1606. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.-H.; Ye, N.; Dong, B. Comparison of Performance between Thermophilic Two-phase and Single-phase Anaerobic Digestion of Dewatered Sludge. Sci. Technol. Eng. 2014, 14, 132–138. (In Chinses) [Google Scholar]
- Liao, X.; Li, H.; Zhang, Y.; Liu, C.; Chen, Q. Accelerated high-solids anaerobic digestion of sewage sludge using low-temperature thermal pretreatment. Int. Biodeterior. Biodegrad. 2016, 106, 141–149. [Google Scholar] [CrossRef]
- Azbar, N.; Speece, R.E. Two-phase two-stage and single stage anaerobic process comparison. J. Environ. Eng. 2001, 127, 240–248. [Google Scholar] [CrossRef]
- Koster, I.W.; Lettinga, G. The influence of ammonium-nitrogen on the specific activity of pelletized methanogenic sludge. Agric. Wastes 1984, 9, 205–216. [Google Scholar] [CrossRef]
- Tandishabo, K.; Nakamura, K.; Umetsu, K.; Takamizawa, K. Distribution and role of Coprothermobacter spp. in anaerobic digesters. J. Biosci. Bioeng. 2012, 114, 518–520. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Hidaka, T.; Tsuno, H. Two-phased hyperthermophilic anaerobic co-digestion of waste activated sludge with kitchen garbage. J. Biosci. Bioeng. 2009, 108, 408–413. [Google Scholar] [CrossRef] [PubMed]
- Pervin, H.M.; Dennis, P.G.; Lim, H.J.; Tyson, G.W.; Batstone, D.J.; Bond, P.L. Drivers of microbial community composition in mesophilic and thermophilic temperature-phased anaerobic digestion pre-treatment reactors. Water Res. 2013, 47, 7098–7108. [Google Scholar] [CrossRef] [PubMed]
- Gagliano, M.C.; Braguglia, C.M.; Gianico, A.; Mininni, G.; Nakamura, K.; Rossetti, S. Thermophilic anaerobic digestion of thermal pretreated sludge: Role of microbial community structure and correlation with process performances. Water Res. 2014, 68, 498–509. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-J.; Wagner, I.; Brice, M.; Kevbrin, V.; Mills, G.; Romanek, C.; Wiegel, J. Thermosediminibacter oceani gen. nov., sp. nov. and Thermosediminibacter litoriperuensis sp. nov., new anaerobic thermophilic bacteria isolated from Peru Margin. Extremophiles 2005, 9, 375–383. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, H.; Wagner, I.D.; Wiegel, J. Caldicoprobacter oshimai gen. nov., sp. nov., an anaerobic, xylanolytic, extremely thermophilic bacterium isolated from sheep faeces, and proposal of Caldicoprobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2010, 60, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Goberna, M.; Insam, H.; Franke-Whittle, I.H. Effect of Biowaste Sludge Maturation on the Diversity of Thermophilic Bacteria and Archaea in an Anaerobic Reactor. Appl. Environ. Microbiol. 2009, 75, 2566–2572. [Google Scholar] [CrossRef] [PubMed]
- Koeck, D.E.; Zverlov, V.V.; Liebl, W.; Schwarz, W.H. Comparative genotyping of Clostridium thermocellum strains isolated from biogas plants: Genetic markers and characterization of cellulolytic potential. Syst. Appl. Microbiol. 2014, 37, 311–319. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.Q.; Ji, P.; Hayashi, J.; Koike, Y.; Wu, X.L.; Kida, K. Characteristic microbial community of a dry thermophilic methanogenic digester: Its long-term stability and change with feeding. Appl. Microbiol. Biotechnol. 2011, 91, 1447–1461. [Google Scholar] [CrossRef] [PubMed]
- Hatamoto, M.; Imachi, H.; Fukayo, S.; Ohashi, A.; Harada, H. Syntrophomonas palmitatica sp. nov., an anaerobic, syntrophic, long-chain fatty-acid-oxidizing bacterium isolated from methanogenic sludge. Int. J. Syst. Evol. Microbiol. 2007, 57, 2137–2142. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cheng, J.J.; Creamer, K.S. Inhibition of anaerobic digestion process: A review. Bioresour. Technol. 2008, 99, 4044–4064. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Roos, S.; Schnurer, A. Syntrophaceticus schinkii gen. nov., sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from a mesophilic anaerobic filter. FEMS Microbiol. Lett. 2010, 309, 100–104. [Google Scholar] [CrossRef] [PubMed]
- Westerholm, M.; Roos, S.; Schnürer, A. Tepidanaerobacter acetatoxydans sp. nov., an anaerobic, syntrophic acetate-oxidizing bacterium isolated from two ammonium-enriched mesophilic methanogenic processes. Syst. Appl. Microbiol. 2011, 34, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Schnürer, A.; Zellner, G.; Svensson, B.H. Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiol. Ecol. 1999, 29, 249–261. [Google Scholar] [CrossRef]
- Karakashev, D.; Batstone, D.J.; Trably, E.; Angelidaki, I. Acetate oxidation is the dominant methanogenic pathway from acetate in the absence of Methanosaetaceae. Appl. Environ. Microbiol. 2006, 72, 5138–5141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, D.P.; Jensen, P.D.; Batstone, D.J. Methanosarcinaceae and acetate-oxidizing pathways dominate in high-rate thermophilic anaerobic digestion of waste-activated sludge. Appl. Environ. Microbiol. 2013, 79, 6491–6500. [Google Scholar] [CrossRef] [PubMed]
- Wett, B.; Takacs, I.; Batstone, D.; Wilson, C.; Murthy, S. Anaerobic model for high-solids or high-temperature digestion—Additional pathway of acetate oxidation. Water Sci. Technol. 2014, 69, 1634–1640. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-H.; Park, J.-H.; Kim, S.-H.; Yu, B.J.; Yoon, J.-J.; Park, H.-D. Evidence of syntrophic acetate oxidation by Spirochaetes during anaerobic methane production. Bioresour. Technol. 2015, 190, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Mazeas, L.; Sghir, A.; Leblon, G.; Bouchez, T. Insights into networks of functional microbes catalysing methanization of cellulose under mesophilic conditions. Environ. Microbiol. 2009, 11, 889–904. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Masse, D.I.; McAllister, T.A.; Beaulieu, C.; Talbot, G.; Kong, Y.; Seviour, R. In situ identification of keratin-hydrolyzing organisms in swine manure inoculated anaerobic digesters. FEMS Microbiol. Ecol. 2011, 78, 451–462. [Google Scholar] [CrossRef] [PubMed]
- Demirel, B.; Scherer, P. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: A review. Rev. Environ. Sci. Bio/Technol. 2008, 7, 173–190. [Google Scholar] [CrossRef]
- Pavlostathis, S.G.; Giraldo-Gomez, E. Kinetics of anaerobic treatment: A critical review. Crit. Rev. Environ. Control 1991, 21, 411–490. [Google Scholar] [CrossRef]


| Parameters | Raw WAS | TPT | TAD | MAD |
|---|---|---|---|---|
| pH | 6.1 ± 0.2 | 6.5 ± 0.0 | 7.8 ± 0.3 | 7.6 ± 0.2 |
| Solid content (%) | 8.75 ± 0.78 | 7.90 ± 0.64 | 6.77 ± 0.40 | 6.23 ± 0.24 |
| VS (%) | 5.64 ± 0.69 | 4.82 ± 0.38 | 3.72 ± 0.22 | 3.21 ± 0.17 |
| VS/TS (%) | 64.98 ± 3.02 | 59.62 ± 2.92 | 55.00 ± 0.45 | 51.61 ± 0.98 |
| SCOD (mg/L) | 6899 ± 585 | 16,747 ± 2190 | 5361 ± 526 | 1661 ± 342 |
| TALK (mg CaO/L) | 479 ± 113 | 1650 ± 423 | 1903 ± 300 | 2476 ± 387 |
| TAN(mg/L) | 522 ± 144 | 1858 ± 163 | 1058 ± 192 | 1245 ± 230 |
| FAN (mg/L) | 0.4 ± 0.3 | 51.4 ± 4.5 | 118.7 ± 21.8 | 42.7 ± 7.9 |
| Clone Library | Bacteria | Archaea | ||||
|---|---|---|---|---|---|---|
| Bac-TPT | Bac-TAD | Bac-MAD | Arc-TPT | Arc-TAD | Arc-MAD | |
| Clone totality | 101 | 101 | 101 | 100 | 74 | 102 |
| Chimera count | 8 | 6 | 9 | 1 | 0 | 0 |
| Valid sequence count | 93 | 95 | 92 | 99 | 74 | 102 |
| OTU count | 23 | 35 | 55 | 9 | 3 | 8 |
| α-diversity indices | ||||||
| Chao1 | 63 | 100 | 325 | 12 | 4 | 23 |
| Shannon index | 2.73 | 3.85 | 4.96 | 0.88 | 0.21 | 0.75 |
| Bacteria | Archaea | ||||||
|---|---|---|---|---|---|---|---|
| Bac-TPT | Bac-TAD | Bac-MAD | Arc-TPT | Arc-TAD | Arc-MAD | ||
| Bac-TPT | 0 | Arc-TPT | 0 | 0.0215 | 0.0228 | ||
| Bac-TAD | 0.3660 | 0 | Arc-TAD | 0 | 0.0092 | ||
| Bac-MAD | 0.5901 | 0.5210 | 0 | Arc-MAD | 0 | ||
| Clone Library | Main Microbes | Relative Abundance | Hydrolysis & Acidogenesis | SAO | Methanogenesis | Unclear | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Protein/Amino Acid | Simple Carbohydrates | Polysaccharide | LCFAs | |||||||
| Bacteria | Bac-TPT | Coprothermobacter | 63.4% | + | ||||||
| Thermosediminibacter | 10.8% | + | + | |||||||
| Caldicoprobacter | 6.5% | + | ||||||||
| Tepidimicrobium | 2.2% | + | + | |||||||
| Bac-TAD | Syntrophaceticus | 4.2% | + | |||||||
| Tepidanaerobacter | 2.1% | + | ||||||||
| Clostridium | 40.0% | + | + | |||||||
| Syntrophomonas | 9.5% | + | ||||||||
| Unclassified 1 | 2.1% | +/− | ||||||||
| Unclassified 2 | 15.8% | + | ||||||||
| Bac-MAD | Mariniphaga | 3.3% | + | |||||||
| Gracilibacter | 2.2% | + | + | |||||||
| Unclassified 1 | 28.3% | +/− | ||||||||
| Unclassified 2 | 15.3% | + | ||||||||
| Archaea | Arc-TAD | Methanothermobacter | 100% | + | ||||||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, J.; Cao, Z.; Hu, Y.; Wang, X.; Wang, G.; Zuo, J.; Wang, K.; Qian, Y. Microbial Insight into a Pilot-Scale Enhanced Two-Stage High-Solid Anaerobic Digestion System Treating Waste Activated Sludge. Int. J. Environ. Res. Public Health 2017, 14, 1483. https://doi.org/10.3390/ijerph14121483
Wu J, Cao Z, Hu Y, Wang X, Wang G, Zuo J, Wang K, Qian Y. Microbial Insight into a Pilot-Scale Enhanced Two-Stage High-Solid Anaerobic Digestion System Treating Waste Activated Sludge. International Journal of Environmental Research and Public Health. 2017; 14(12):1483. https://doi.org/10.3390/ijerph14121483
Chicago/Turabian StyleWu, Jing, Zhiping Cao, Yuying Hu, Xiaolu Wang, Guangqi Wang, Jiane Zuo, Kaijun Wang, and Yi Qian. 2017. "Microbial Insight into a Pilot-Scale Enhanced Two-Stage High-Solid Anaerobic Digestion System Treating Waste Activated Sludge" International Journal of Environmental Research and Public Health 14, no. 12: 1483. https://doi.org/10.3390/ijerph14121483






