Oral Microcosm Biofilms Grown under Conditions Progressing from Peri-Implant Health, Peri-Implant Mucositis, and Peri-Implantitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Selection of Donors
2.2. Microcosm Biofilm Model
2.3. Harvesting of Microcosm Biofilms
2.4. Characterisation by Culture Media Analyses
2.5. Characterisation by 16S rRNA Amplification and Sequencing
2.6. Imaging Microcosm Biofilms
2.7. Sequencing Data Analysis
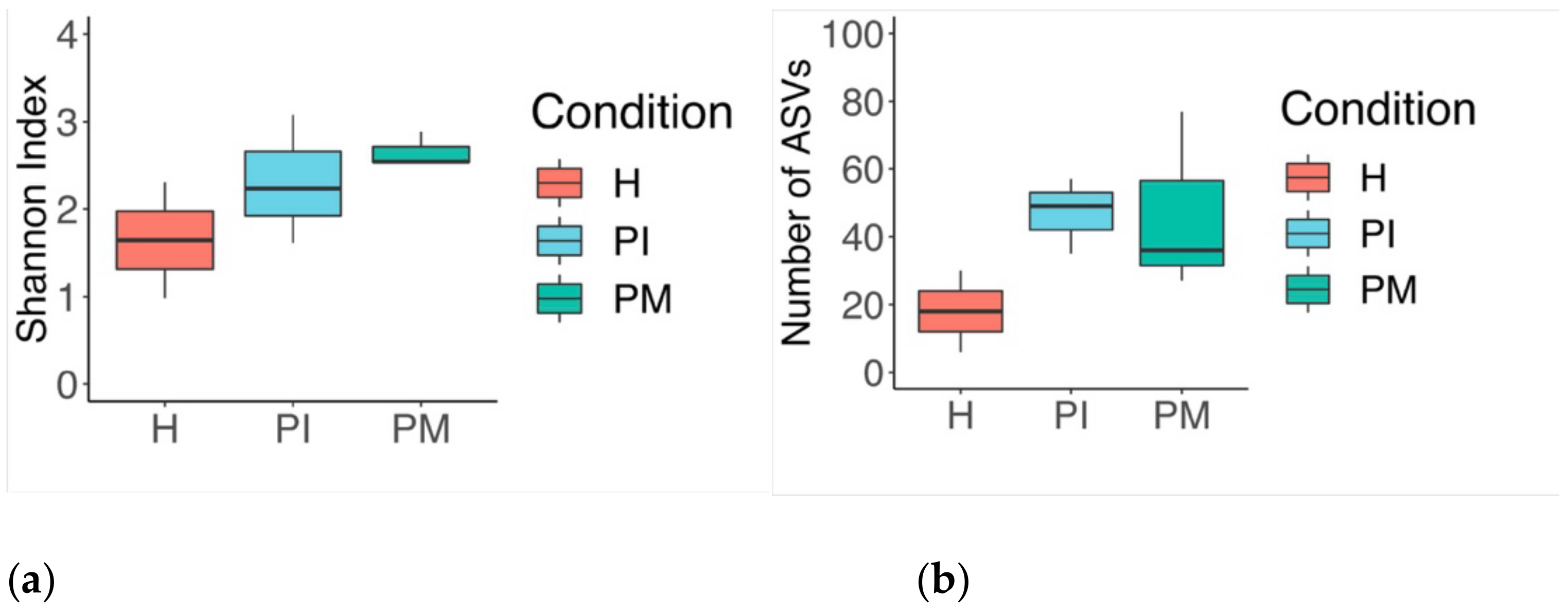
3. Results
3.1. Culture Media Analysis
3.2. Analysis of the 16S rRNA Gene and Comparative Sequencing
3.2.1. Phylum
3.2.2. Genus
3.2.3. Sub-Analyses at the Species Level
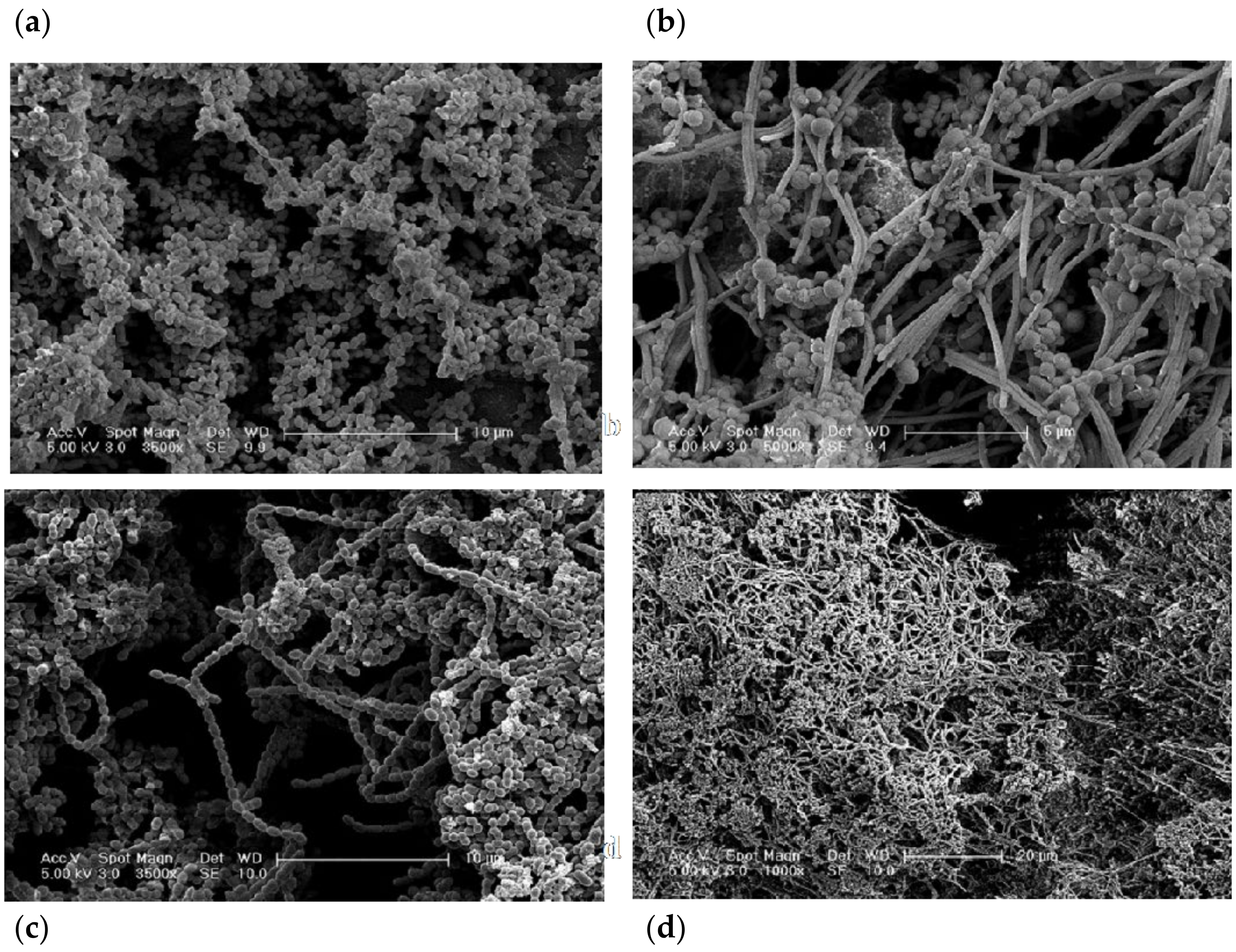
3.3. Structural Analysis
4. Discussion
4.1. Resemblance of the Experiments
4.2. Methods to Analyse and Characterise the Microcosm Biofilms
4.3. CDFF Modelling
4.4. Characterisation of Biofilm Architecture
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elani, H.W.; Starr, J.R.; Da Silva, J.D.; Gallucci, G.O. Trends in Dental Implant Use in the U.S., 1999–2016, and Projections to 2026. J. Dent. Res. 2018, 97, 1424–1430. [Google Scholar] [CrossRef]
- Roccuzzo, A.; Stähli, A.; Monje, A.; Sculean, A.; Salvi, G.E. Peri-Implantitis: A Clinical Update on Prevalence and Surgical Treatment Outcomes. J. Clin. Med. 2021, 10, 1107. [Google Scholar] [CrossRef] [PubMed]
- Kordbacheh Changi, K.; Finkelstein, J.; Papapanou, P.N. Peri-implantitis prevalence, incidence rate, and risk factors: A study of electronic health records at a U.S. dental school. Clin. Oral Implants Res. 2019, 30, 306–314. [Google Scholar] [CrossRef] [PubMed]
- Berglundh, T.; Armitage, G.; Araujo, M.G.; Avila-Ortiz, G.; Blanco, J.; Camargo, P.M.; Chen, S.; Cochran, D.; Derks, J.; Figuero, E.; et al. Peri-implant diseases and conditions: Consensus report of workgroup 4 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 2018, 45 (Suppl. 20), S286–S291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belibasakis, G.N.; Manoil, D. Microbial Community-Driven Etiopathogenesis of Peri-Implantitis. J. Dent. Res. 2021, 100, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, K.; Derks, J.; Wennström, J.L.; Petzold, M.; Berglundh, T. Health economic aspects of implant-supported restorative therapy. Clin. Oral Implants Res. 2022, 33, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Ammann, T.W.; Bostanci, N.; Belibasakis, G.N.; Thurnheer, T. Validation of a quantitative real-time PCR assay and comparison with fluorescence microscopy and selective agar plate counting for species-specific quantification of an in vitro subgingival biofilm model. J. Periodont. Res. 2013, 48, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Kommerein, N.; Doll, K.; Stumpp, N.S.; Stiesch, M. Development and characterization of an oral multispecies biofilm implant flow chamber model. PLoS ONE 2018, 13, e0196967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cieplik, F.; Zaura, E.; Brandt, B.W.; Buijs, M.J.; Buchalla, W.; Crielaard, W.; Laine, M.L.; Deng, D.M.; Exterkate, R.A. Microcosm biofilms cultured from different oral niches in periodontitis patients. J. Oral Microbiol. 2018, 11, 1551596. [Google Scholar] [CrossRef]
- Hope, C.K.; Bakht, K.; Burnside, G.; Martin, G.C.; Burnett, G.; de Josselin de Jong, E.; Higham, S.M. Reducing the variability between constant-depth film fermenter experiments when modelling oral biofilm. J. Appl. Microbiol. 2012, 113, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Coulter, J.; Jakubovics, N.S.; Preshaw, P.M.; German, M.J. An in vitro model to assess effects of a desensitising agent on bacterial biofilm formation. Acta Biomater. Odontol. Scand. 2019, 5, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, M.C.; Alonso-Español, A.; Ribeiro-Vidal, H.; Alonso, B.; Herrera, D.; Sanz, M. Relevance of Biofilm Models in Periodontal Research: From Static to Dynamic Systems. Microorganisms 2021, 9, 428. [Google Scholar] [CrossRef] [PubMed]
- Benli, M.; Petit, C.; Tenenbaum, H.; Huck, O. In vitro Assessment of Peri-implantitis Treatment Procedures: A Review. Open Dent. J. 2019, 13, 267–273. [Google Scholar] [CrossRef]
- Sousa, V.; Mardas, N.; Spratt, D.; Hassan, I.A.; Walters, N.J.; Beltrán, V.; Donos, N. The Effect of Microcosm Biofilm Decontamination on Surface Topography, Chemistry, and Biocompatibility Dynamics of Implant Titanium Surfaces. Int. J. Mol. Sci. 2022, 23, 10033. [Google Scholar] [CrossRef]
- Wilson, M. Lethal photosensitisation of oral bacteria and its potential application in the photodynamic therapy of oral infections. Photochem. Photobiol. Sci. 2004, 3, 412–418. [Google Scholar] [CrossRef] [PubMed]
- Blank, E.; Grischke, J.; Winkel, A.; Eberhard, J.; Kommerein, N.; Doll, K.; Yang, I.; Stiesch, M. Evaluation of biofilm colonization on multi-part dental implants in a rat model. BMC Oral Health 2021, 21, 313. [Google Scholar] [CrossRef]
- Sissons, C.H.; Cutress, T.W.; Hoffman, M.P.; Wakefield, J.S. A multi-station dental plaque microcosm (artificial mouth) for the study of plaque growth, metabolism, pH, and mineralization. J. Dent. Res. 1991, 70, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.; Sissons, C. A comparison of human dental plaque microcosm biofilms grown in an undefined medium and a chemically defined artificial saliva. Arch. Oral Biol. 2001, 46, 477–486. [Google Scholar] [CrossRef]
- Hope, C.K.; Wilson, M. Measuring the thickness of an outer layer of viable bacteria in an oral biofilm by viability mapping. J. Microbiol. Methods 2003, 54, 403–410. [Google Scholar] [CrossRef]
- Navazesh, M.; Christensen, C.M. A comparison of whole mouth resting and stimulated salivary measurement procedures. J. Dent. Res. 1982, 61, 1158–1162. [Google Scholar] [CrossRef]
- Pratten, J. Growing oral biofilms in a constant depth film fermentor (CDFF). Curr. Protoc. Microbiol. 2007, 6, 1B.5. [Google Scholar] [CrossRef] [PubMed]
- Dalwai, F.; Spratt, D.A.; Pratten, J. Modeling shifts in microbial populations associated with health or disease. Appl. Environ. Microbiol. 2006, 72, 3678–3684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uitto, V.-J. Gingival crevice fluid—An introduction. Periodontology 2000 2003, 31, 9–11. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, D.J.; Marsh, P.D.; Allison, C.; Schilling, K.M. Effect of oxygen, inoculum composition and flow rate on development of mixed-culture oral biofilms. Microbiology 1996, 142, 623–629. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Author Correction: Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 1091. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Lozupone, C.; Lladser, M.E.; Knights, D.; Stombaugh, J.; Knight, R. UniFrac: An effective distance metric for microbial community comparison. ISME J. 2010, 5, 169–172. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Wilson, M. Use of constant depth film fermentor in studies of biofilms of oral bacteria. Methods Enzymol. 1999, 310, 264–279. [Google Scholar] [CrossRef]
- Pratten, J.; Wilson, M. Antimicrobial susceptibility and composition of microcosm dental plaques supplemented with sucrose. Antimicrob. Agents Chemother. 1999, 43, 1595–1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rüdiger, S.G.; Carlén, A.; Meurman, J.H.; Kari, K.; Olsson, J. Dental biofilms at healthy and inflamed gingival margins. J. Clin. Periodontol. 2002, 29, 524–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, C.; Coulter, W.A. Continuous monitoring of pH and Eh in bacterial plaque grown on a tooth in an artificial mouth. Appl. Microbiol. 1975, 29, 141–144. [Google Scholar] [CrossRef] [PubMed]
- Goodson, J.M. Gingival crevice fluid flow. Periodontology 2000 2003, 31, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Cimasoni, G. Crevicular fluid updated. Monogr. Oral Sci. 1983, 12, 1–152. [Google Scholar]
- Apse, P.; Ellen, R.P.; Overall, C.M.; Zarb, G.A. Microbiota and crevicular fluid collagenase activity in the osseointegrated dental implant sulcus: A comparison of sites in edentulous and partially edentulous patients. J. Periodontal. Res. 1989, 24, 96–105. [Google Scholar] [CrossRef]
- Kinniment, S.L.; Wimpenny, J.W.; Adams, D.; Marsh, P.D. Development of a steady-state oral microbial biofilm community using the constant-depth film fermenter. Microbiology 1996, 142, 631–638. [Google Scholar] [CrossRef] [Green Version]
- Pratten, J.; Wilson, M.; Spratt, D.A. Characterization of in vitro oral bacterial biofilms by traditional and molecular methods. Oral. Microbiol. Immunol. 2003, 18, 45–49. [Google Scholar] [CrossRef]
- Polge, C.; Smith, A.U.; Parkes, A.S. Revival of spermatozoa after vitrification and dehydration at low temperatures. Nature 1949, 164, 666. [Google Scholar] [CrossRef]
- Thurnheer, T.; Gmür, R.; Guggenheim, B. Multiplex FISH analysis of a six-species bacterial biofilm. J. Microbiol. Methods 2004, 56, 37–47. [Google Scholar] [CrossRef]
- Mombelli, A.; Décaillet, F. The characteristics of biofilms in peri-implant disease. J. Clin. Periodontol. 2011, 38 (Suppl. 11), 203–213. [Google Scholar] [CrossRef] [PubMed]
- Pratten, J.; Barnett, P.; Wilson, M. Composition and susceptibility to chlorhexidine of multispecies biofilms of oral bacteria. Appl. Environ. Microbiol. 1998, 64, 3515–3519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McBain, A.J.; Bartolo, R.G.; Catrenich, C.E.; Charbonneau, D.; Ledder, R.G.; Gilbert, P. Effects of triclosan-containing rinse on the dynamics and antimicrobial susceptibility of in vitro plaque ecosystems. Antimicrob. Agents Chemother. 2003, 47, 3531–3538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marsh, P.D. Microbial ecology of dental plaque and its significance in health and disease. Adv. Dent. Res. 1994, 8, 263–271. [Google Scholar] [CrossRef]
- Kumar, P.S.; Mason, M.R.; Brooker, M.R.; O’Brien, K. Pyrosequencing reveals unique microbial signatures associated with healthy and failing dental implants. J. Clin. Periodontol. 2012, 39, 425–433. [Google Scholar] [CrossRef] [Green Version]
- Dabdoub, S.M.; Tsigarida, A.A.; Kumar, P.S. Patient-specific analysis of periodontal and peri-implant microbiomes. J. Dent. Res. 2013, 92, 168S–175S. [Google Scholar] [CrossRef] [Green Version]
- Koyanagi, T.; Sakamoto, M.; Takeuchi, Y.; Ohkuma, M.; Izumi, Y. Analysis of microbiota associated with peri-implantitis using 16S rRNA gene clone library. J. Oral Microbiol. 2010, 2, 1–7. [Google Scholar] [CrossRef]
- Al-Ahmad, A.; Muzafferiy, F.; Anderson, A.C.; Wölber, J.P.; Ratka-Krüger, P.; Fretwurst, T.; Nelson, K.; Vach, K.; Hellwig, E. Shift of microbial composition of peri-implantitis-associated oral biofilm as revealed by 16S rRNA gene cloning. J. Med. Microbiol. 2018, 67, 332–340. [Google Scholar] [CrossRef]
- Tsigarida, A.A.; Dabdoub, S.M.; Nagaraja, H.N.; Kumar, P.S. The Influence of Smoking on the Peri-Implant Microbiome. J. Dent. Res. 2015, 94, 1202–1217. [Google Scholar] [CrossRef] [Green Version]
- Sousa, V.; Nibali, L.; Spratt, D.; Dopico, J.; Mardas, N.; Petrie, A.; Donos, N. Peri-implant and periodontal microbiome diversity in aggressive periodontitis patients: A pilot study. Clin. Oral Implants Res. 2017, 28, 558–570. [Google Scholar] [CrossRef]
- Nibali, L.; Sousa, V.; Davrandi, M.; Spratt, D.; Alyahya, Q.; Dopico, J.; Donos, N. Differences in the periodontal microbiome of successfully treated and persistent aggressive periodontitis. J. Clin. Periodontol. 2020, 47, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Al-Radha, A.S.D.; Dymock, D.; Younes, C.; O’Sullivan, D. Surface properties of titanium and zirconia dental implant materials and their effect on bacterial adhesion. J. Dent. 2012, 40, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Aruni, W.; Chioma, O.; Fletcher, H.M. Filifactor alocis: The Newly Discovered Kid on the Block with Special Talents. J. Dent. Res. 2014, 93, 725–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charalampakis, G.; Rabe, P.; Leonhardt, Å.; Dahlén, G. A follow-up study of peri-implantitis cases after treatment. J. Clin. Periodontol. 2011, 38, 864–871. [Google Scholar] [CrossRef] [PubMed]
- Botero, J.E.; González, A.M.; Mercado, R.A.; Olave, G.; Contreras, A. Subgingival microbiota in peri-implant mucosa lesions and adjacent teeth in partially edentulous patients. J. Periodontol. 2005, 76, 1490–1495. [Google Scholar] [CrossRef] [PubMed]
- Nowzari, H.; Botero, J.E.; DeGiacomo, M.; Villacres, M.C.; Rich, S.K. Microbiology and cytokine levels around healthy dental implants and teeth. Clin. Implant Dent. Relat. Res. 2008, 10, 166–173. [Google Scholar] [CrossRef]
- Rams, T.E.; Degener, J.E.; van Winkelhoff, A.J. Antibiotic resistance in human peri-implantitis microbiota. Clin. Oral Implants Res. 2014, 25, 82–90. [Google Scholar] [CrossRef] [Green Version]
- Wu, M.; Li, X. Klebsiella pneumoniae and Pseudomonas aeruginosa. In Molecular Medical Microbiology, 2nd ed.; Academic Press: Boston, MA, USA, 2015; pp. 1547–1564. [Google Scholar]
- Sanz, M.; Newman, M.G.; Nachnani, S.; Holt, R.; Stewart, R.; Flemmig, T. Characterization of the subgingival microbial flora around endosteal sapphire dental implants in partially edentulous patients. Int. J. Oral Maxillofac. Implants 1990, 5, 247–253. [Google Scholar]
- Silverstein, L.H.; Kurtzman, D.; Garnick, J.J.; Schuster, G.S.; Steflik, D.E.; Moskowitz, M.E. The microbiota of the peri-implant region in health and disease. Implant Dent. 1994, 3, 170–174. [Google Scholar] [CrossRef] [PubMed]
- Pontoriero, R.; Tonelli, M.P.; Carnevale, G.; Mombelli, A.; Nyman, S.R.; Lang, N.P. Experimentally induced peri-implant mucositis. A clinical study in Humans. Clin. Oral. Implants Res. 1994, 5, 254–259. [Google Scholar] [CrossRef]
- Leonhardt, A.; Renvert, S.; Dahlén, G. Microbial findings at failing implants. Clin. Oral Implants Res. 1999, 10, 339–345. [Google Scholar] [CrossRef]
- Stokman, M.A.; van Winkelhoff, A.J.; Vissink, A.; Spijkervet, F.K.L.; Raghoebar, G.M. Bacterial colonization of the peri-implant sulcus in dentate patients: A prospective observational study. Clin. Oral Investig. 2017, 21, 717–724. [Google Scholar] [CrossRef]
- Charalampakis, G.; Ramberg, P.; Dahlén, G.; Berglundh, T.; Abrahamsson, I. Effect of cleansing of biofilm formed on titanium discs. Clin. Oral Implants Res. 2015, 26, 931–936. [Google Scholar] [CrossRef] [PubMed]
- Costerton, J.W. Overview of microbial biofilms. J. Ind. Microbiol. 1995, 15, 137–140. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sousa, V.; Spratt, D.; Davrandi, M.; Mardas, N.; Beltrán, V.; Donos, N. Oral Microcosm Biofilms Grown under Conditions Progressing from Peri-Implant Health, Peri-Implant Mucositis, and Peri-Implantitis. Int. J. Environ. Res. Public Health 2022, 19, 14088. https://doi.org/10.3390/ijerph192114088
Sousa V, Spratt D, Davrandi M, Mardas N, Beltrán V, Donos N. Oral Microcosm Biofilms Grown under Conditions Progressing from Peri-Implant Health, Peri-Implant Mucositis, and Peri-Implantitis. International Journal of Environmental Research and Public Health. 2022; 19(21):14088. https://doi.org/10.3390/ijerph192114088
Chicago/Turabian StyleSousa, Vanessa, Dave Spratt, Mehmet Davrandi, Nikos Mardas, Víctor Beltrán, and Nikolaos Donos. 2022. "Oral Microcosm Biofilms Grown under Conditions Progressing from Peri-Implant Health, Peri-Implant Mucositis, and Peri-Implantitis" International Journal of Environmental Research and Public Health 19, no. 21: 14088. https://doi.org/10.3390/ijerph192114088




