Performance and Microbial Community Dynamics in Anaerobic Digestion of Waste Activated Sludge: Impact of Immigration
Abstract
:1. Introduction
2. Material and Methods
2.1. Collection of Sludge
2.2. Bioreactor Operation
2.3. Sampling and Analyses
2.4. DNA Extraction and High-Throughput Sequencing
2.5. Real-Time PCR
2.6. Statistical Analyses and Estimation of Net Growth Rates of the Sequencing Data
3. Results
3.1. Bioreactor Operation
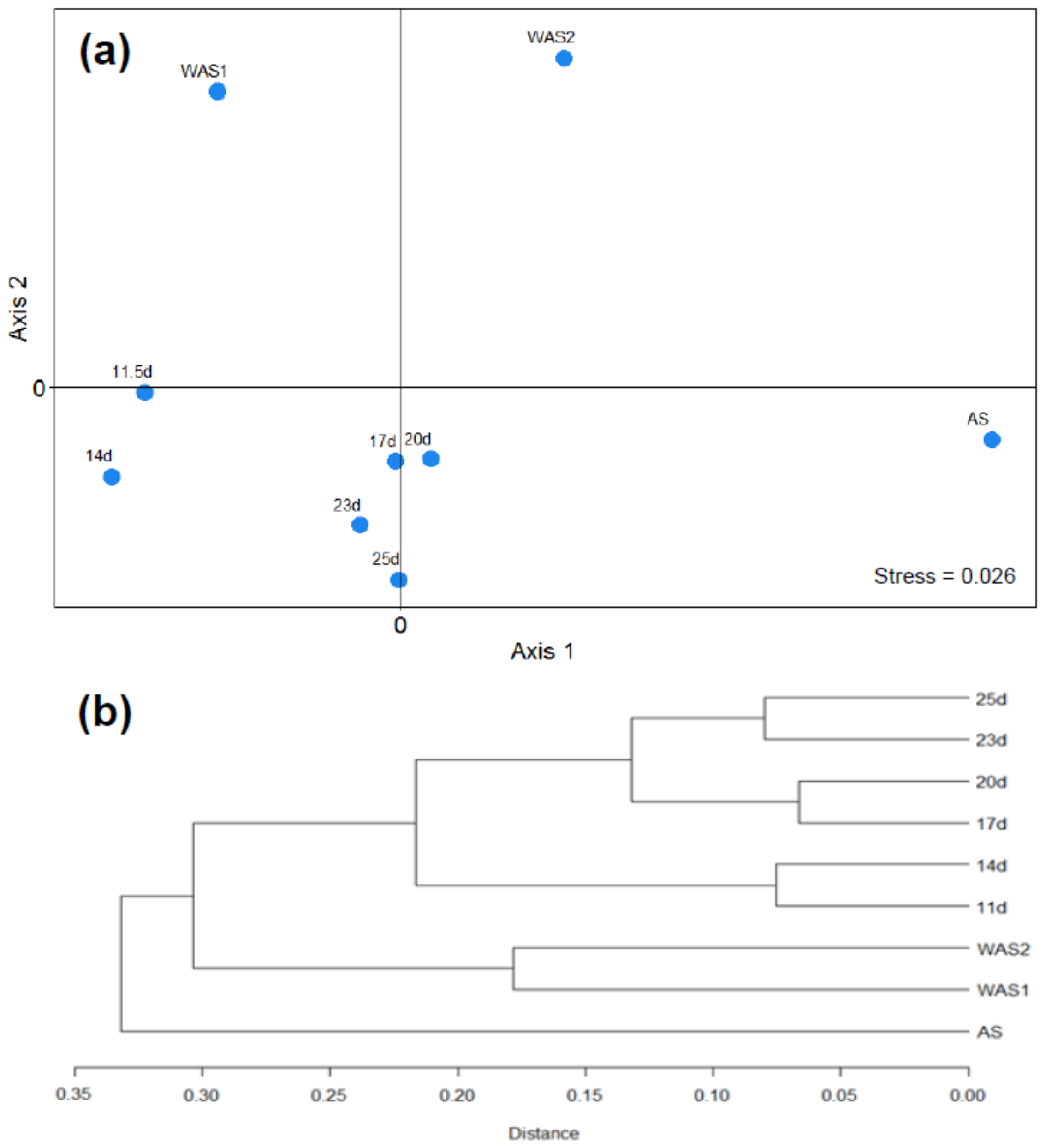
3.2. Microbial Community Results
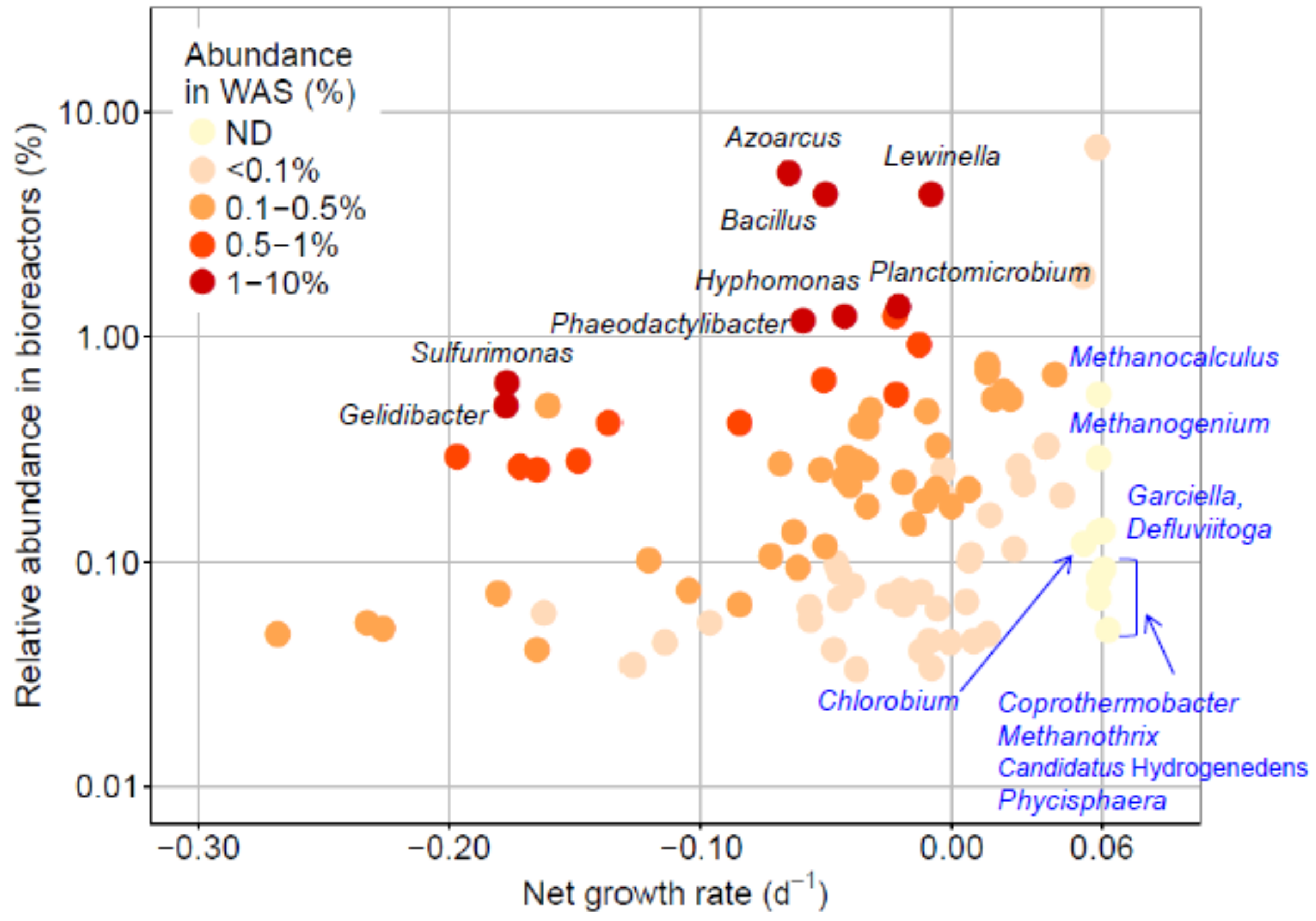
3.3. Estimation of Net Growth Rates
3.4. Relationship between Process Parameters and Abundance of Microbial Taxa
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
References
- McCarty, P.L.; Bae, J.; Kim, J. Domestic wastewater treatment as a net energy producer—Can this be achieved? Environ. Sci. Technol. 2011, 45, 7100–7106. [Google Scholar] [CrossRef] [PubMed]
- Oladejo, J.; Shi, K.; Luo, X.; Yang, G.; Wu, T. A review of sludge-to-energy recovery methods. Energies 2018, 12, 60. [Google Scholar] [CrossRef]
- Kobayashi, T.; Li, Y.Y.; Harada, H. Analysis of microbial community structure and diversity in the thermophilic anaerobic digestion of waste activated sludge. Water Sci. Technol. 2008, 57, 1199–1205. [Google Scholar] [CrossRef] [PubMed]
- Karakashev, D.; Batstone, D.J.; Angelidaki, I. Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors. Appl. Environ. Microbiol. 2005, 71, 331–338. [Google Scholar] [CrossRef] [PubMed]
- Saunders, A.M.; Albertsen, M.; Vollertsen, J.; Nielsen, P.H. The activated sludge ecosystem contains a core community of abundant organisms. ISME J. 2016, 10, 11–20. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.G.; Lee, S.; Lee, C.; Hwang, K.; Hwang, S. Qualitative and quantitative assessment of microbial community in batch anaerobic digestion of secondary sludge. Bioresour. Technol. 2010, 101, 9461–9470. [Google Scholar] [CrossRef]
- Zhou, B.W.; Shin, S.G.; Hwang, K.; Ahn, J.-H.; Hwang, S. Effect of microwave irradiation on cellular disintegration of Gram positive and negative cells. Appl. Microbiol. Biotechnol. 2010, 87, 765–770. [Google Scholar] [CrossRef]
- De Vrieze, J.; Raport, L.; Roume, H.; Vilchez-Vargas, R.; Jáuregui, R.; Pieper, D.H.; Boon, N. The full-scale anaerobic digestion microbiome is represented by specific marker populations. Water Res. 2016, 104, 101–110. [Google Scholar] [CrossRef]
- Connelly, S.; Shin, S.G.; Dillon, R.J.; Ijaz, U.Z.; Quince, C.; Sloan, W.T.; Collins, G. Bioreactor scalability: Laboratory-scale bioreactor design influences performance, ecology, and community physiology in expanded granular sludge bed bioreactors. Front. Microbiol. 2017, 8, 664. [Google Scholar] [CrossRef]
- Müller, B.; Sun, L.; Westerholm, M.; Schnürer, A. Bacterial community composition and fhs profiles of low- and high-ammonia biogas digesters reveal novel syntrophic acetate-oxidising bacteria. Biotechnol. Biofuels 2016, 9, 1–18. [Google Scholar] [CrossRef]
- Lee, J.; Kim, E.; Han, G.; Tongco, J.V.; Shin, S.G.; Hwang, S. Microbial communities underpinning mesophilic anaerobic digesters treating food wastewater or sewage sludge: A full-scale study. Bioresour. Technol. 2018, 259, 388–397. [Google Scholar] [CrossRef]
- Černý, M.; Vítězová, M.; Vítěz, T.; Bartoš, M.; Kushkevych, I. Variation in the distribution of hydrogen producers from the Clostridiales order in biogas reactors depending on different input substrates. Energies 2018, 11, 3270. [Google Scholar] [CrossRef]
- Shin, S.G.; Koo, T.; Lee, J.; Han, G.; Cho, K.; Kim, W.; Hwang, S. Correlations between bacterial populations and process parameters in four full-scale anaerobic digesters treating sewage sludge. Bioresour. Technol. 2016, 214, 711–721. [Google Scholar] [CrossRef]
- Meng, X.; Zhang, Y.; Sui, Q.; Zhang, J.; Wang, R.; Yu, D.; Wang, Y.; Wei, Y. Biochemical conversion and microbial community in response to ternary pH buffer system during anaerobic digestion of swine manure. Energies 2018, 11, 2991. [Google Scholar] [CrossRef]
- Wacławek, S.; Grübel, K.; Silvestri, D.; Padil, V.V.T.; Wacławek, M.; Černík, M.; Varma, R.S. Disintegration of wastewater activated sludge (WAS) for improved biogas production. Energies 2018, 12, 21. [Google Scholar] [CrossRef]
- Al-Addous, M.; Saidan, M.N.; Bdour, M.; Alnaief, M. Evaluation of biogas production from the co-digestion of municipal food waste and wastewater sludge at refugee camps using an automated methane potential test system. Energies 2018, 12, 32. [Google Scholar] [CrossRef]
- Eaton, A.D.; Clesceri, L.S.; Greenberg, A.E. Standard Methods for the Examination of Water and Wastewater, 21st ed.; American Public Health Association: Washington, DC, USA, 2005; pp. 2–56. ISBN ISBN 978-0875530475. [Google Scholar]
- Dubois, M.; Gilles, K.A.; Hamilton, J.K.; Rebers, P.A.; Smith, F. Colorimetric determination of sugars and related substances. Anal. Chem. 1956, 28, 350–356. [Google Scholar] [CrossRef]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef]
- Cho, K.; Shin, S.G.; Lee, J.; Koo, T.; Kim, W.; Hwang, S. Nitrification resilience and community dynamics of ammonia-oxidizing bacteria with respect to ammonia loading shock in a nitrification reactor treating steel wastewater. J. Biosci. Bioeng. 2016, 122, 196–202. [Google Scholar] [CrossRef]
- Cho, K.; Jeong, Y.; Seo, K.W.; Lee, S.; Smith, A.L.; Shin, S.G.; Cho, S.-K.; Park, C. Effects of changes in temperature on treatment performance and energy recovery at mainstream anaerobic ceramic membrane bioreactor for food waste recycling wastewater treatment. Bioresour. Technol. 2018, 256, 137–144. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Meth. 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Doi, T.; Matsumoto, H.; Abe, J.; Morita, S. Feasibility study on the application of rhizosphere microflora of rice for the biohydrogen production from wasted bread. Int. J. Hydrogen Energ. 2009, 34, 1735–1743. [Google Scholar] [CrossRef]
- Yu, Y.; Lee, C.; Kim, J.; Hwang, S. Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol. Bioeng. 2005, 89, 670–679. [Google Scholar] [CrossRef]
- Koo, T.; Shin, S.G.; Lee, J.; Han, G.; Kim, W.; Cho, K.; Hwang, S. Identifying methanogen community structures and their correlations with performance parameters in four full-scale anaerobic sludge digesters. Bioresour. Technol. 2017, 228, 368–373. [Google Scholar] [CrossRef]
- Lee, C.; Kim, J.; Chinalia, F.; Shin, S.G.; Hwang, S. Unusual bacterial populations observed in a full-scale municipal sludge digester affected by intermittent seawater inputs. J. Ind. Microbiol. Biotechnol. 2009, 36, 769–773. [Google Scholar] [CrossRef]
- Madigan, M.T.; Martinko, J.M. Brock Biology of Microorganisms, 11th ed.; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2006; p. 42. ISBN ISBN 978-0131443297. [Google Scholar]
- Ahn, J.-H.; Shin, S.G.; Hwang, S. Effect of microwave irradiation on the disintegration and acidogenesis of municipal secondary sludge. Chem. Eng. J. 2009, 153, 145–150. [Google Scholar] [CrossRef]
- Carrère, H.; Antonopoulou, G.; Affes, R.; Passos, F.; Battimelli, A.; Lyberatos, G.; Ferrer, I. Review of feedstock pretreatment strategies for improved anaerobic digestion: From lab-scale research to full-scale application. Bioresour. Technol. 2016, 199, 386–397. [Google Scholar] [CrossRef]
- Kim, J.; Park, C.; Kim, T.-H.; Lee, M.; Kim, S.; Kim, S.-W.; Lee, J. Effects of various pretreatments for enhanced anaerobic digestion with waste activated sludge. J. Biosci. Bioeng. 2003, 95, 271–275. [Google Scholar] [CrossRef]
- Zhang, Y.; Feng, Y.; Yu, Q.; Xu, Z.; Quan, X. Enhanced high-solids anaerobic digestion of waste activated sludge by the addition of scrap iron. Bioresour. Technol. 2014, 159, 297–304. [Google Scholar] [CrossRef]
- De la Rubia, M.A.; Perez, M.; Romero, L.I.; Sales, D. Effect of solids retention time (SRT) on pilot scale anaerobic thermophilic sludge digestion. Process Biochem. 2006, 41, 79–86. [Google Scholar] [CrossRef]
- Moset, V.; Poulsen, M.; Wahid, R.; Højberg, O.; Møller, H.B. Mesophilic versus thermophilic anaerobic digestion of cattle manure: Methane productivity and microbial ecology. Microb. Biotechnol. 2015, 8, 787–800. [Google Scholar] [CrossRef]
- Mao, C.; Feng, Y.; Wang, X.; Ren, G. Review on research achievements of biogas from anaerobic digestion. Renew. Sustain. Energy Rev. 2015, 45, 540–555. [Google Scholar] [CrossRef]
- Shin, S.G.; Han, G.; Lim, J.; Lee, C.; Hwang, S. A comprehensive microbial insight into two-stage anaerobic digestion of food waste-recycling wastewater. Water Res. 2010, 44, 4838–4849. [Google Scholar] [CrossRef]
- Miron, Y.; Zeeman, G.; van Lier, J.B.; Lettinga, G. The role of sludge retention time in the hydrolysis and acidification of lipids, carbohydrates and proteins during digestion of primary sludge in CSTR systems. Water Res. 2000, 34, 1705–1713. [Google Scholar] [CrossRef]
- Siegrist, H.; Vogt, D.; Garcia-Heras, J.L.; Gujer, W. Mathematical model for meso- and thermophilic anaerobic sewage sludge digestion. Environ. Sci. Technol. 2002, 36, 1113–1123. [Google Scholar] [CrossRef]
- Bolzonella, D.; Pavan, P.; Battistoni, P.; Cecchi, F. Mesophilic anaerobic digestion of waste activated sludge: Influence of the solid retention time in the wastewater treatment process. Process Biochem. 2005, 40, 1453–1460. [Google Scholar] [CrossRef]
- Hidaka, T.; Wang, F.; Tsumori, J. Comparative evaluation of anaerobic digestion for sewage sludge and various organic wastes with simple modeling. Waste Manag. 2015, 43, 144–151. [Google Scholar] [CrossRef]
- Guo, J.; Peng, Y.; Ni, B.-J.; Han, X.; Fan, L.; Yuan, Z. Dissecting microbial community structure and methane-producing pathways of a full-scale anaerobic reactor digesting activated sludge from wastewater treatment by metagenomic sequencing. Microb. Cell Factories 2015, 14, 33. [Google Scholar] [CrossRef]
- Han, G.; Shin, S.G.; Lee, J.; Shin, J.; Hwang, S. A comparative study on the process efficiencies and microbial community structures of six full-scale wet and semi-dry anaerobic digesters treating food wastes. Bioresour. Technol. 2017, 245, 869–875. [Google Scholar] [CrossRef]
- Goux, X.; Calusinska, M.; Fossépré, M.; Benizri, E.; Delfosse, P. Start-up phase of an anaerobic full-scale farm reactor – Appearance of mesophilic anaerobic conditions and establishment of the methanogenic microbial community. Bioresour. Technol. 2016, 212, 217–226. [Google Scholar] [CrossRef]
- Gonzalez-Martinez, A.; Rodriguez-Sanchez, A.; Lotti, T.; Garcia-Ruiz, M.-J.; Osorio, F.; Gonzalez-Lopez, J.; van Loosdrecht, M.C.M. Comparison of bacterial communities of conventional and A-stage activated sludge systems. Sci. Rep. 2016, 6, 18786. [Google Scholar] [CrossRef]
- Stams, A.J.; Sousa, D.Z.; Kleerebezem, R.; Plugge, C.M. Role of syntrophic microbial communities in high-rate methanogenic bioreactors. Water Sci. Technol. 2012, 66, 352–362. [Google Scholar] [CrossRef]
- Rivière, D.; Desvignes, V.; Pelletier, E.; Chaussonnerie, S.; Guermazi, S.; Weissenbach, J.; Li, T.; Camacho, P.; Sghir, A. Towards the definition of a core of microorganisms involved in anaerobic digestion of sludge. ISME J. 2009, 3, 700–714. [Google Scholar] [CrossRef]
- Jumas-Bilak, E.; Roudière, L.; Marchandin, H. Description of ‘Synergistetes’ phyl. nov. and emended description of the phylum ‘Deferribacteres’ and of the family Syntrophomonadaceae, phylum ‘Firmicutes’. Int. J. Syst. Evol. Microbiol. 2009, 59, 1028–1035. [Google Scholar] [CrossRef]
- Li, J.; Rui, J.; Yao, M.; Zhang, S.; Yan, X.; Wang, Y.; Yan, Z.; Li, X. Substrate type and free ammonia determine bacterial community structure in full-scale mesophilic anaerobic digesters treating cattle or swine manure. Front. Microbiol. 2015, 6, 1337. [Google Scholar] [CrossRef]
- Nesbø, C.; Bradnan, D.; Adebusuyi, A.; Dlutek, M.; Petrus, A.; Foght, J.; Doolittle, W.F.; Noll, K. Mesotoga prima gen. nov., sp. nov., the first described mesophilic species of the Thermotogales. Extremophiles 2012, 16, 387–393. [Google Scholar] [CrossRef]
- Wang, P.; Yu, Z.; Zhao, J.; Zhang, H. Do microbial communities in an anaerobic bioreactor change with continuous feeding sludge into a full-scale anaerobic digestion system? Bioresour. Technol. 2018, 249, 89–98. [Google Scholar] [CrossRef]
- Khan, S.T.; Fukunaga, Y.; Nakagawa, Y.; Harayama, S. Emended descriptions of the genus Lewinella and of Lewinella cohaerens, Lewinella nigricans and Lewinella persica, and description of Lewinella lutea sp. nov. and Lewinella marina sp. nov. Int. J. Syst. Evol. Microbiol. 2007, 57, 2946–2951. [Google Scholar] [CrossRef]
- Inagaki, F.; Takai, K.; Nealson, K.H.; Horikoshi, K. Sulfurovum lithotrophicum gen. nov., sp. nov., a novel sulfur-oxidizing chemolithoautotroph within the ε-Proteobacteria isolated from Okinawa Trough hydrothermal sediments. Int. J. Syst. Evol. Microbiol. 2004, 54, 1477–1482. [Google Scholar] [CrossRef]
- Röling, W.F.M.; van Breukelen, B.M.; Braster, M.; Lin, B.; van Verseveld, H.W. Relationships between microbial community structure and hydrochemistry in a landfill leachate-polluted aquifer. Appl. Environ. Microbiol. 2001, 67, 4619–4629. [Google Scholar] [CrossRef]
- Kulichevskaya, I.S.; Ivanova, A.A.; Detkova, E.N.; Rijpstra, W.I.C.; Sinninghe Damsté, J.S.; Dedysh, S.N. Planctomicrobium piriforme gen. nov., sp. nov., a stalked planctomycete from a littoral wetland of a boreal lake. Int. J. Syst. Evol. Microbiol. 2015, 65, 1659–1665. [Google Scholar] [CrossRef]
- Kim, H.-S.; Bang, J.J.; Lee, S.-S. Gelidibacter flavus sp. nov., Isolated from Activated Sludge of Seawater Treatment System. Curr. Microbiol. 2017, 74, 1247–1252. [Google Scholar] [CrossRef]
- Mountfort, D.O.; Brulla, W.J.; Krumholz, L.R.; Bryant, M.P. Syntrophus buswellii gen. nov., sp. nov.: A benzoate catabolizer from methanogenic ecosystems. Int. J. Syst. Evol. Microbiol. 1984, 34, 216–217. [Google Scholar] [CrossRef]
- Li, Y.; Sun, Y.; Li, L.; Yuan, Z. Acclimation of acid-tolerant methanogenic propionate-utilizing culture and microbial community dissecting. Bioresour. Technol. 2018, 250, 117–123. [Google Scholar] [CrossRef]
- Gagliano, M.C.; Braguglia, C.M.; Petruccioli, M.; Rossetti, S. Ecology and biotechnological potential of the thermophilic fermentative Coprothermobacter spp. FEMS Microbiol. Ecol. 2015. [Google Scholar] [CrossRef]
- Wu, G.; Zheng, D.; Xing, L. Nitritation and N2O emission in a denitrification and nitrification two-sludge system treating high ammonium containing wastewater. Water 2014, 6, 2978. [Google Scholar] [CrossRef]
- Wu, L.; Yang, Y.; Chen, S.; Jason Shi, Z.; Zhao, M.; Zhu, Z.; Yang, S.; Qu, Y.; Ma, Q.; He, Z.; Zhou, J.; He, Q. Microbial functional trait of rRNA operon copy numbers increases with organic levels in anaerobic digesters. ISME J. 2017, 11, 2874. [Google Scholar] [CrossRef]
- Kim, J.; Kim, W.; Lee, C. Absolute dominance of hydrogenotrophic methanogens in full-scale anaerobic sewage sludge digesters. J. Environ. Sci. 2013, 25, 2272–2280. [Google Scholar] [CrossRef]
- Woodcock, S.; Sloan, W.T. Biofilm community succession: A neutral perspective. Microbiology 2017, 163, 664–668. [Google Scholar] [CrossRef]
- Liu, Z.; Cichocki, N.; Hübschmann, T.; Süring, C.; Ofiţeru, I.D.; Sloan, W.T.; Grimm, V.; Müller, S. Neutral mechanisms and niche differentiation in steady-state insular microbial communities revealed by single cell analysis. Environ. Microbiol. 2019, 21, 164–181. [Google Scholar] [CrossRef]
- Cho, S.-K.; Kim, D.-H.; Quince, C.; Im, W.-T.; Oh, S.-E.; Shin, S.G. Low-strength ultrasonication positively affects methanogenic granules toward higher AD performance: Implications from microbial community shift. Ultrason. Sonochem. 2016, 32, 198–203. [Google Scholar] [CrossRef] [PubMed]




| Parameter | Unit | Value | Standard Deviation |
|---|---|---|---|
| pH | – | 6.9 | 0.3 |
| Chemical oxygen demand (COD) | mg/L | 15,189 | 227 |
| Soluble COD (SCOD) | mg/L | 107 | 118 |
| Total solids (TS) | mg/L | 23,032 | 568 |
| Volatile solids (VS) | mg/L | 10,173 | 202 |
| Total suspended solids (TSS) | mg/L | 13,575 | 260 |
| Volatile suspended solids (VSS) | mg/L | 8930 | 42 |
| Crude carbohydrate | mg/L | 2056 | 304 |
| Crude protein | mg/L | 5478 | 31 |
| Crude lipid | mg/L | 429 | 35 |
| Volatile fatty acids (VFAs) | mg/L | 86.6 | 3.5 |
| Ethanol | mg/L | ND | ND |
| Total ammonia nitrogen (TAN) | mg/L | 24.2 | 2.0 |
| Sodium (Na+) | mg/L | 2619 | 2 |
| Chloride (Cl-) | mg/L | 5595 | 3 |
| Group | Genus | HRT | COD Removal | MPRa | MYb |
|---|---|---|---|---|---|
| I | Methanogenium | 0.83 * | 0.83 * | −0.83 * | 0.66 |
| Acetobacteroides | 0.89 ** | 0.89 ** | −0.89 ** | 0.94 ** | |
| Mesotoga | 0.83 * | 0.83 * | −0.83 * | 0.94 ** | |
| Thermovirga | 0.94 ** | 0.94 ** | −0.94 ** | 1.00 *** | |
| Variovorax | 0.83 * | 0.83 * | −0.83 * | 0.77 | |
| Aquabacterium | 1.00 *** | 1.00 *** | −1.00 *** | 0.94 ** | |
| Soehngenia | 0.94 ** | 0.94 ** | −0.94 ** | 1.00 *** | |
| Arcobacter | 0.94 ** | 0.94 ** | −0.94 ** | 1.00 *** | |
| Fusibacter | 0.94 ** | 0.94 ** | −0.94 ** | 0.83* | |
| Spirochaeta | 0.83 * | 0.83 * | −0.83 * | 0.94 ** | |
| II | Gelidibacter | −0.89 ** | −0.89 ** | 0.89 ** | −0.71 |
| Chondromyces | −0.94 ** | −0.94 ** | 0.94 ** | −0.83 * | |
| Phaeodactylibacter | −0.83 * | −0.83 * | 0.83 * | −0.89 ** | |
| Clostridium | −0.89 ** | −0.89 ** | 0.89 ** | −0.77 | |
| Mycobacterium | −0.89 ** | −0.89 ** | 0.89 ** | −0.83 * | |
| Methanocalculus | −0.83 * | −0.83 * | 0.83 * | −0.77 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.; Cho, S.-K.; Lee, J.; Hwang, K.; Chung, J.W.; Jang, H.-N.; Shin, S.G. Performance and Microbial Community Dynamics in Anaerobic Digestion of Waste Activated Sludge: Impact of Immigration. Energies 2019, 12, 573. https://doi.org/10.3390/en12030573
Shin J, Cho S-K, Lee J, Hwang K, Chung JW, Jang H-N, Shin SG. Performance and Microbial Community Dynamics in Anaerobic Digestion of Waste Activated Sludge: Impact of Immigration. Energies. 2019; 12(3):573. https://doi.org/10.3390/en12030573
Chicago/Turabian StyleShin, Juhee, Si-Kyung Cho, Joonyeob Lee, Kwanghyun Hwang, Jae Woo Chung, Hae-Nam Jang, and Seung Gu Shin. 2019. "Performance and Microbial Community Dynamics in Anaerobic Digestion of Waste Activated Sludge: Impact of Immigration" Energies 12, no. 3: 573. https://doi.org/10.3390/en12030573





