Bioconversion Pathway of CO2 in the Presence of Ethanol by Methanogenic Enrichments from Production Water of a High-Temperature Petroleum Reservoir
Abstract
:1. Introduction
2. Materials and Methods
2.1. Enrichment Cultures
2.2. Chemical Analysis
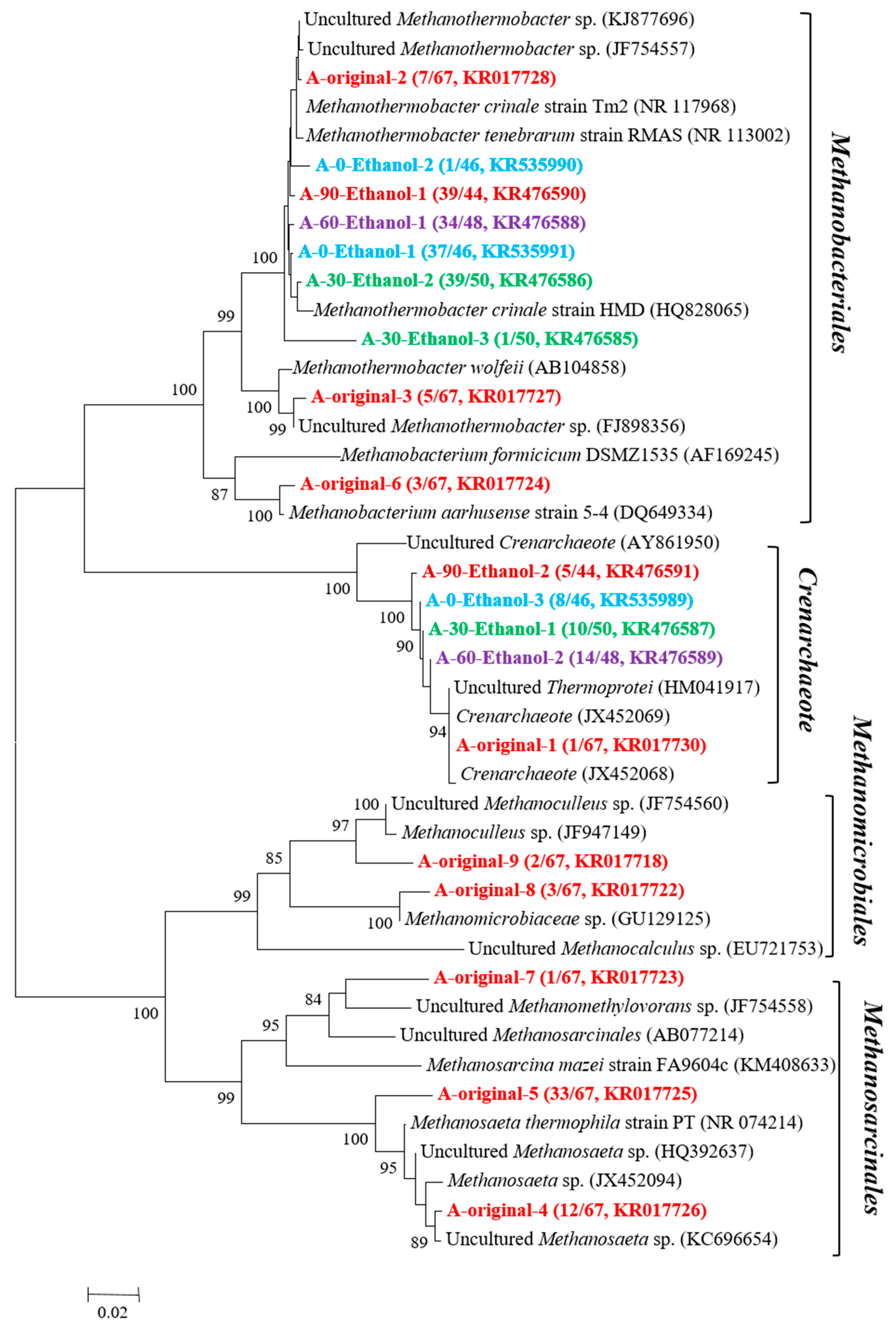
2.3. DNA Extraction and Construction of Bacterial and Archaeal 16S rRNA Gene Clone Libraries
2.4. Phylogenetic Analysis
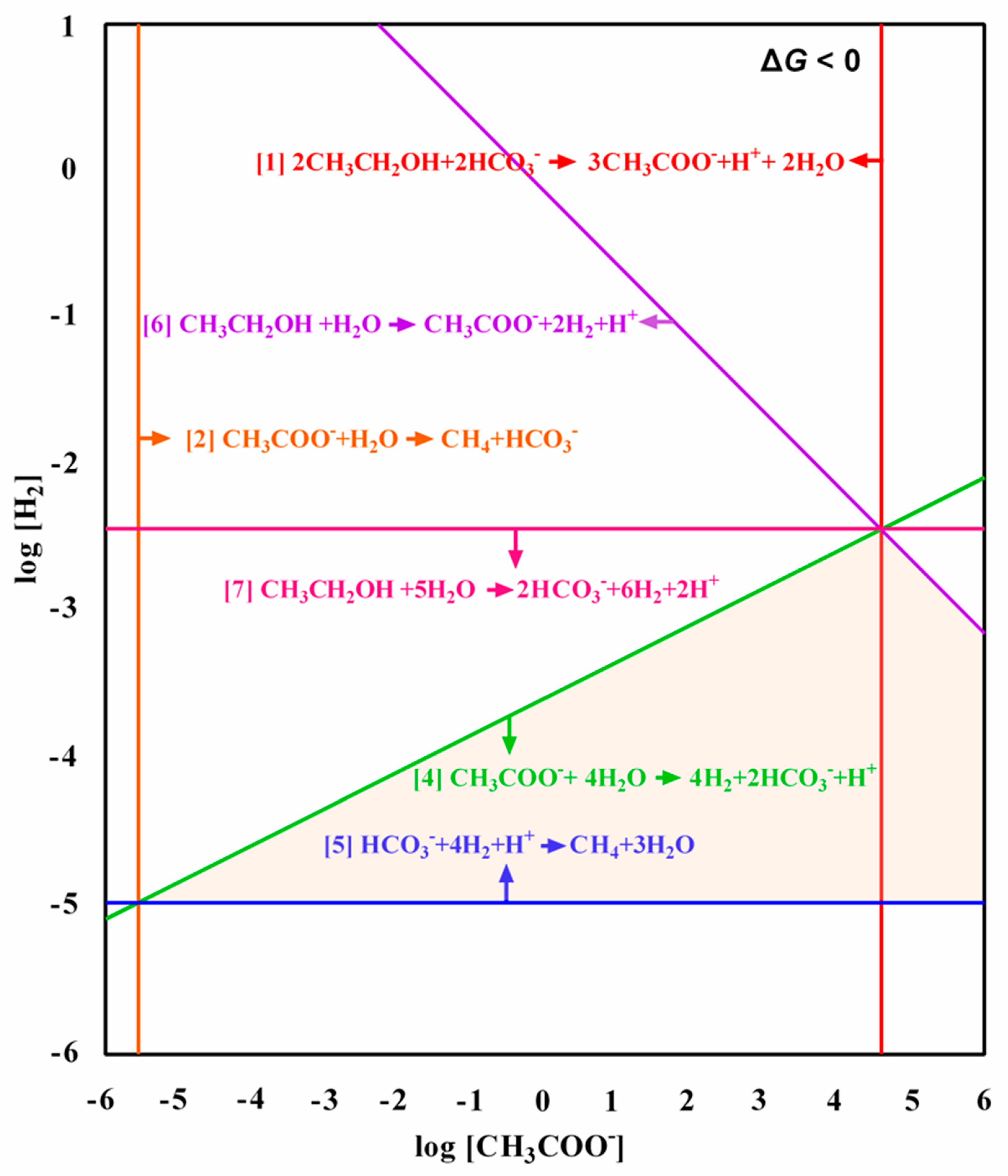
2.5. Thermodynamics Analysis
3. Results and Discussion
3.1. Biotransformation of CO2 into Methane in the Presence of Ethanol
3.2. Microorganisms Occurring in the Enrichment Cultures
3.3. Metabolic Pathways for Methanogenesis from Ethanol
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Appendix A
| Pathways | ΔG°25°C (kJ/mol) | ΔG°55°C (kJ/mol) | ΔG°′55°C (kJ/mol) | |
|---|---|---|---|---|
| Route 1 Bioconversion of ethanol to acetate, linked to acetoclastic methanogenesis | ||||
| 2CH3CH2OH + 2HCO3− → 3CH3COO− + H++ 2H2O 3CH3COO− + 3H2O → 3CH4 + 3HCO3− SUM: 2CH3CH2OH + H2O → 3CH4 + HCO3− + H+ | −45.87 −93.45 −139.32 | −42.58 −104.61 −147.19 | −86.56 −104.61 −191.17 | [1] [2] [3] |
| Route 2 Bioconversion of ethanol to acetate, linked to syntrophic acetate oxidation and CO2-reducing methanogenesis | ||||
| 2CH3CH2OH + 2HCO3− → 3CH3COO− + H+ + 2H2O 3CH3COO− + 12H2O → 12H2 + 6HCO3− + 3H+ 3HCO3− + 12H2 + 3H+→ 3CH4 + 9H2O SUM: 2CH3CH2OH + H2O → 3CH4 + HCO3− + H+ | −45.87 432.51 −525.96 −139.32 | −42.58 401.88 −506.49 −147.19 | −86.56 269.94 −374.55 −191.17 | [1] [4] [5] [3] |
| Route 3 Bioconversion of ethanol to hydrogen and acetate, linked to acetoclastic methanogenesis and CO2-reducing methanogenesis | ||||
| 2CH3CH2OH + 2H2O → 2CH3COO− + 4H2 + 2H+ 2CH3COO− + 2H2O → 2CH4 + 2HCO3− HCO3− + 4H2 + H+ → CH4 + 3H2O SUM: 2CH3CH2OH + H2O → 3CH4 + HCO3− + H+ | 98.30 −62.30 −175.32 −139.32 | 91.38 −69.74 −168.83 −147.19 | 3.42 −69.74 −124.85 −191.17 | [6] [2] [5] [3] |
| Route 4 Bioconversion of ethanol to hydrogen and acetate, linked to syntrophic acetate oxidation and CO2-reducing methanogenesis | ||||
| 2CH3CH2OH + 2H2O → 2CH3COO− + 4H2 + 2H+ 2CH3COO− + 8H2O → 8H2 + 4HCO3− + 2H+ 3HCO3− + 12H2 + 3H+ → 3CH4 + 9H2O SUM: 2CH3CH2OH + H2O → 3CH4 + HCO3− + H+ | 98.30 288.34 −525.96 −139.32 | 91.38 267.92 −506.49 −147.19 | 3.421 79.96 −374.55 −191.17 | [6] [4] [5] [3] |
| Route 5 Bioconversion of ethanol to hydrogen and CO2, linked to CO2-reducing methanogenesis | ||||
| 2CH3CH2OH + 10H2O → 4HCO3− + 12H2 + 4H+ 3HCO3− + 12H2 + 3H+ → 3CH4 + 9H2O SUM: 2CH3CH2OH + H2O → 3CH4 + HCO3− + H+ | 386.64 −525.96 −139.32 | 359.30 −506.49 −147.19 | 183.36 −374.55 −191.17 | [7] [5] [3] |
References
- Benson, S.M.; Orr, F.M. Carbon dioxide capture and storage. MRS Bull. 2011, 33, 303–305. [Google Scholar] [CrossRef]
- Haszeldine, R.S. Carbon capture and storage: How green can black be? Science 2009, 325, 1647–1652. [Google Scholar] [CrossRef] [PubMed]
- Leung, D.Y.C.; Caramanna, G.; Maroto-Valer, M.M. An overview of current status of carbon dioxide capture and storage technologies. Renew. Sustain. Energy Rev. 2014, 39, 426–443. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Zhao, N.; Wei, W.; Sun, Y. A review of research progress on CO2 capture, storage, and utilization in Chinese Academy of Sciences. Fuel 2013, 108, 112–130. [Google Scholar] [CrossRef]
- Hallmann, C.; Schwark, L.; Grice, K. Community dynamics of anaerobic bacteria in deep petroleum reservoirs. Nat. Geosci. 2008, 1, 588. [Google Scholar] [CrossRef]
- Mbadinga, S.M.; Wang, L.-Y.; Zhou, L.; Liu, J.-F.; Gu, J.-D.; Mu, B.-Z. Microbial communities involved in anaerobic degradation of alkanes. Int. Biodeterior. Biodegrad. 2011, 65, 1–13. [Google Scholar] [CrossRef]
- Jones, D.M.; Head, I.M.; Gray, N.D.; Adams, J.J.; Rowan, A.K.; Aitken, C.M.; Bennett, B.; Huang, H.; Brown, A.; Bowler, B.F.J.; et al. Crude-oil biodegradation via methanogenesis in subsurface petroleum reservoirs. Nature 2007, 451, 176. [Google Scholar] [CrossRef]
- Orphan, V.J.; Taylor, L.T.; Hafenbradl, D.; Delong, E.F. Culture-dependent and culture-independent characterization of microbial assemblages associated with high-temperature petroleum reservoirs. Appl. Environ. Microbiol. 2000, 66, 700–711. [Google Scholar] [CrossRef]
- Grabowski, A.; Nercessian, O.; Fayolle, F.; Blanchet, D.; Jeanthon, C. Microbial diversity in production waters of a low-temperature biodegraded oil reservoir. FEMS Microbiol. Ecol. 2005, 54, 427–443. [Google Scholar] [CrossRef]
- Aramaki, N.; Tamamura, S.; Ueno, A.; Badrul, A.A.K.M.; Murakami, T.; Tamazawa, S.; Yamaguchi, S.; Aoyama, H.; Kaneko, K. Experimental investigation on the feasibility of industrial methane production in the subsurface environment via microbial activities in northern Hokkaido, Japan – A process involving subsurface cultivation and gasification. Energy Convers. Manag. 2017, 153, 566–575. [Google Scholar] [CrossRef]
- Bhatia, S.K.; Kim, S.-H.; Yoon, J.-J.; Yang, Y.-H. Current status and strategies for second generation biofuel production using microbial systems. Energy Convers. Manag. 2017, 148, 1142–1156. [Google Scholar] [CrossRef]
- Dolfing, J.; Larter, S.R.; Head, I.M. Thermodynamic constraints on methanogenic crude oil biodegradation. ISME J. 2008, 2, 442. [Google Scholar] [CrossRef] [PubMed]
- Gieg, L.M.; Duncan, K.E.; Suflita, J.M. Bioenergy production via microbial conversion of residual oil to natural gas. Appl. Environ. Microbiol. 2008, 74, 3022–3029. [Google Scholar] [CrossRef] [PubMed]
- Aitken, C.M.; Jones, D.M.; Larter, S.R. Anaerobic hydrocarbon biodegradation in deep subsurface oil reservoirs. Nature 2004, 431, 291. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Liang, B.; Wang, L.-Y.; Zhou, L.; Mbadinga, S.M.; Gu, J.-D.; Mu, B.-Z. Microbial reduction of CO2 from injected NaH13CO3 with degradation of n-hexadecane in the enrichment culture derived from a petroleum reservoir. Int. Biodeterior. Biodegrad. 2018, 127, 192–200. [Google Scholar] [CrossRef]
- Berk, H.; Thauer, R.K. Function of coenzyme F420-dependent NADP reductase in methanogenic archaea containing an NADP-dependent alcohol dehydrogenase. Arch. Microbiol. 1997, 168, 396–402. [Google Scholar] [CrossRef]
- Fardeau, M.L.; Magot, M.; Patel, B.K.; Thomas, P.; Garcia, J.L.; Ollivier, B. Thermoanaerobacter subterraneus sp. nov., a novel thermophile isolated from oilfield water. Int. J. Syst. Evol. Microbiol. 2000, 50, 2141–2149. [Google Scholar] [CrossRef]
- Yang, G.-C.; Zhou, L.; Mbadinga, S.M.; Liu, J.-F.; Yang, S.-Z.; Gu, J.-D.; Mu, B.-Z. Formate-dependent microbial conversion of CO2 and the dominant pathways of methanogenesis in production water of high-temperature oil reservoirs amended with bicarbonate. Front. Microbiol. 2016, 7, 365. [Google Scholar] [CrossRef]
- Wang, L.-Y.; Li, W.; Mbadinga, S.M.; Liu, J.-F.; Gu, J.-D.; Mu, B.-Z. Methanogenic microbial community composition of oily sludge and its enrichment amended with alkanes incubated for over 500 days. Geomicrobiol. J. 2012, 29, 716–726. [Google Scholar] [CrossRef]
- Lv, L.; Mbadinga, S.M.; Wang, L.-Y.; Liu, J.-F.; Gu, J.-D.; Mu, B.-Z.; Yang, S.-Z. Acetoclastic methanogenesis is likely the dominant biochemical pathway of palmitate degradation in the presence of sulfate. Appl. Microbiol. Biotechnol. 2015, 99, 7757–7769. [Google Scholar] [CrossRef]
- Conrad, R. Quantification of methanogenic pathways using stable carbon isotopic signatures: A review and a proposal. Org. Geochem. 2005, 36, 739–752. [Google Scholar] [CrossRef]
- Mbadinga, S.M.; Li, K.-P.; Zhou, L.; Wang, L.-Y.; Yang, S.-Z.; Liu, J.-F.; Gu, J.-D.; Mu, B.-Z. Analysis of alkane-dependent methanogenic community derived from production water of a high-temperature petroleum reservoir. Appl. Microbiol. Biotechnol. 2012, 96, 531–542. [Google Scholar] [CrossRef] [PubMed]
- Barton, H.A.; Taylor, M.R.; Pace, N.R. Molecular phylogenetic analysis of a bacterial community in an oligotrophic cave environment. Geomicrobiol. J. 2004, 21, 11–20. [Google Scholar] [CrossRef]
- Gantner, S.; Andersson, A.F.; Alonso-Sáez, L.; Bertilsson, S. Novel primers for 16S rRNA-based archaeal community analyses in environmental samples. J. Microbiol. Methods 2011, 84, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Amend, J.P.; Shock, E.L. Energetics of overall metabolic reactions of thermophilic and hyperthermophilic Archaea and Bacteria. FEMS Microbiol. Rev. 2001, 25, 175–243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, L.; Zhou, L.; Mbadinga, S.M.; Gu, J.-D.; Mu, B.-Z. Accelerated CO2 reduction to methane for energy by zero valent iron in oil reservoir production waters. Energy 2018, 147, 663–671. [Google Scholar] [CrossRef]
- Kawaguchi, H.; Sakuma, T.; Nakata, Y.; Kobayashi, H.; Endo, K.; Sato, K. Methane production by Methanothermobacter thermautotrophicus to recover energy from carbon dioxide sequestered in geological reservoirs. J. Biosci. Bioeng. 2010, 110, 106–108. [Google Scholar] [CrossRef]
- Radianingtyas, H.; Wright, P.C. Alcohol dehydrogenases from thermophilic and hyperthermophilic archaea and bacteria. FEMS Microbiol. Rev. 2003, 27, 593–616. [Google Scholar] [CrossRef] [Green Version]
- Head, I.; Gray, N.; Aitken, C.; Sherry, A.; Jones, M.; Larter, S. Hydrocarbon activation under sulfate-reducing and methanogenic conditions proceeds by different mechanisms. In Proceedings of the EGU General Assembly Conference, Vienna, Austria, 2–7 May 2010. [Google Scholar]
- Liu, Y.; Whitman, W.B. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann. N. Y. Acad. Sci. 2008, 1125, 171–189. [Google Scholar] [CrossRef]
- Demirel, B.; Scherer, P. The roles of acetotrophic and hydrogenotrophic methanogens during anaerobic conversion of biomass to methane: A review. Rev. Environ. Sci. Bio-Technol. 2008, 7, 173–190. [Google Scholar] [CrossRef]
- Bryant, M.P.; Wolin, E.A.; Wolin, M.J.; Wolfe, R.S. Methanobacillus omelianskii, a symbiotic association of two species of bacteria. Arch. Mikrobiol. 1967, 59, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.-C.; Zhou, L.; Mbadinga, S.M.; You, J.; Yang, H.-Z.; Liu, J.-F.; Yang, S.-Z.; Gu, J.-D.; Mu, B.-Z. Activation of CO2-reducing methanogens in oil reservoir after addition of nutrient. J. Biosci. Bioeng. 2016, 122, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.-K.; Li, G.-Q.; Zhao, L.-X.; Dai, X.-C.; Tian, H.-M.; Dai, L.-B.; Wang, H.-B.; Huang, H.-D.; Chen, Y.-H.; Ma, T. Dynamic processes of indigenous microorganisms from a low-temperature petroleum reservoir during nutrient stimulation. J. Biosci. Bioeng. 2014, 117, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Sato, K.; Kawaguchi, H.; Kobayashi, H. Bio-electrochemical conversion of carbon dioxide to methane in geological storage reservoirs. Energy Convers. Manag. 2013, 66, 343–350. [Google Scholar] [CrossRef]
- Barker, H.A. Studies upon the methane fermentation. IV. The isolation and culture of Methanobacterium Omelianskii. Antonie Leeuwenhoek 1939, 6, 201–220. [Google Scholar] [CrossRef]
- Tatton, M.J.; Archer, D.B.; Powell, G.E.; Parker, M.L. Methanogenesis from ethanol by defined mixed continuous cultures. Appl. Environ. Microbiol. 1989, 55, 440–445. [Google Scholar]
- Abbasi, T.; Tauseef, S.M.; Abbasi, S.A. Biogas Energy; Springer: New York, NY, USA, 2012; pp. 25–34. [Google Scholar]
- Yamane, K.; Maki, H.; Nakayama, T.; Nakajima, T.; Nomura, N.; Uchiyama, H.; Kitaoka, M. Diversity and similarity of microbial communities in petroleum crude oils produced in Asia. Biosci. Biotechnol. Biochem. 2008, 72, 2831–2839. [Google Scholar] [CrossRef] [PubMed]
- Silva, T.R.; Verde, L.C.L.; Santos Neto, E.V.; Oliveira, V.M. Diversity analyses of microbial communities in petroleum samples from Brazilian oil fields. Int. Biodeterior. Biodegrad. 2013, 81, 57–70. [Google Scholar] [CrossRef]
- Pham, V.D.; Hnatow, L.L.; Zhang, S.; Fallon, R.D.; Jackson, S.C.; Tomb, J.F.; DeLong, E.F.; Keeler, S.J. Characterizing microbial diversity in production water from an Alaskan mesothermic petroleum reservoir with two independent molecular methods. Environ. Microbiol. 2009, 11, 176–187. [Google Scholar] [CrossRef]
- Kobayashi, H.; Kawaguchi, H.; Endo, K.; Mayumi, D.; Sakata, S.; Ikarashi, M.; Miyagawa, Y.; Maeda, H.; Sato, K. Analysis of methane production by microorganisms indigenous to a depleted oil reservoir for application in Microbial Enhanced Oil Recovery. J. Biosci. Bioeng. 2012, 113, 84–87. [Google Scholar] [CrossRef]
- Gieg, L.M.; Davidova, I.A.; Duncan, K.E.; Suflita, J.M. Methanogenesis, sulfate reduction and crude oil biodegradation in hot Alaskan oilfields. Environ. Microbiol. 2010, 12, 3074–3086. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Y.-L.; Hanada, S.; Kamagata, Y.; Guo, R.-B.; Sekiguchi, Y. Lactivibrio alcoholicus gen. nov., sp. nov., an anaerobic, mesophilic, lactate-, alcohol-, carbohydrate- and amino-acid-degrading bacterium in the phylum Synergistetes. Int. J. Syst. Evol. Microbiol. 2014, 64, 2137–2145. [Google Scholar] [CrossRef] [PubMed]
- Maune, M.W.; Tanner, R.S. Description of Anaerobaculum hydrogeniformans sp. nov., an anaerobe that produces hydrogen from glucose, and emended description of the genus Anaerobaculum. Int. J. Syst. Evol. Microbiol. 2012, 62, 832–838. [Google Scholar] [CrossRef] [PubMed]
- Sekiguchi, Y.; Muramatsu, M.; Imachi, H.; Narihiro, T.; Ohashi, A.; Harada, H.; Hanada, S.; Kamagata, Y. Thermodesulfovibrio aggregans sp. nov. and Thermodesulfovibrio thiophilus sp. nov., anaerobic, thermophilic, sulfate-reducing bacteria isolated from thermophilic methanogenic sludge, and emended description of the genus Thermodesulfovibrio. Int. J. Syst. Evol. Microbiol. 2008, 58, 2541–2548. [Google Scholar] [CrossRef] [PubMed]
- Hui, L.; Shi-Zhong, Y.; Bo-Zhong, M.; Zhao-Feng, R.; Jie, Z. Molecular phylogenetic diversity of the microbial community associated with a high-temperature petroleum reservoir at an offshore oilfield. FEMS Microbiol. Ecol. 2007, 60, 74–84. [Google Scholar] [Green Version]
- Nazina, T.N.; Shestakova, N.M.; Grigor’yan, A.A.; Mikhailova, E.M.; Tourova, T.P.; Poltaraus, A.B.; Feng, C.; Ni, F.; Belyaev, S.S. Phylogenetic diversity and activity of anaerobic microorganisms of high-temperature horizons of the Dagang oil field (P.R. China). Microbiology 2006, 75, 55–65. [Google Scholar] [CrossRef]
- Cheng, L.; Dai, L.; Li, X.; Zhang, H.; Lu, Y. Isolation and characterization of Methanothermobacter crinale sp nov., a novel hydrogenotrophic methanogen from the shengli oil field. Appl. Environ. Microbiol. 2011, 77, 5212–5219. [Google Scholar] [CrossRef] [PubMed]
- Daisuke, M.; Hanako, M.; Hideyoshi, Y.; Susumu, S.; Haruo, M.; Yoshihiro, M.; Masayuki, I.; Mio, T.; Yoichi, K. Evidence for syntrophic acetate oxidation coupled to hydrogenotrophic methanogenesis in the high-temperature petroleum reservoir of Yabase oil field (Japan). Environ. Microbiol. 2011, 13, 1995–2006. [Google Scholar]
- Berdugoclavijo, C.; Gieg, L.M. Conversion of crude oil to methane by a microbial consortium enriched from oil reservoir production waters. Front. Microbiol. 2014, 5, 197. [Google Scholar]
- Mayumi, D.; Dolfing, J.; Sakata, S.; Maeda, H.; Miyagawa, Y.; Ikarashi, M.; Tamaki, H.; Takeuchi, M.; Nakatsu, C.H.; Kamagata, Y. Carbon dioxide concentration dictates alternative methanogenic pathways in oil reservoirs. Nat. Commun. 2013, 4, 1998. [Google Scholar] [CrossRef] [Green Version]
- Lv, Z.; Hu, M.; Harms, H.; Richnow, H.H.; Liebetrau, J.; Nikolausz, M. Stable isotope composition of biogas allows early warning of complete process failure as a result of ammonia inhibition in anaerobic digesters. Bioresour. Technol. 2014, 167, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Schink, B. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 1997, 61, 262–280. [Google Scholar] [PubMed]
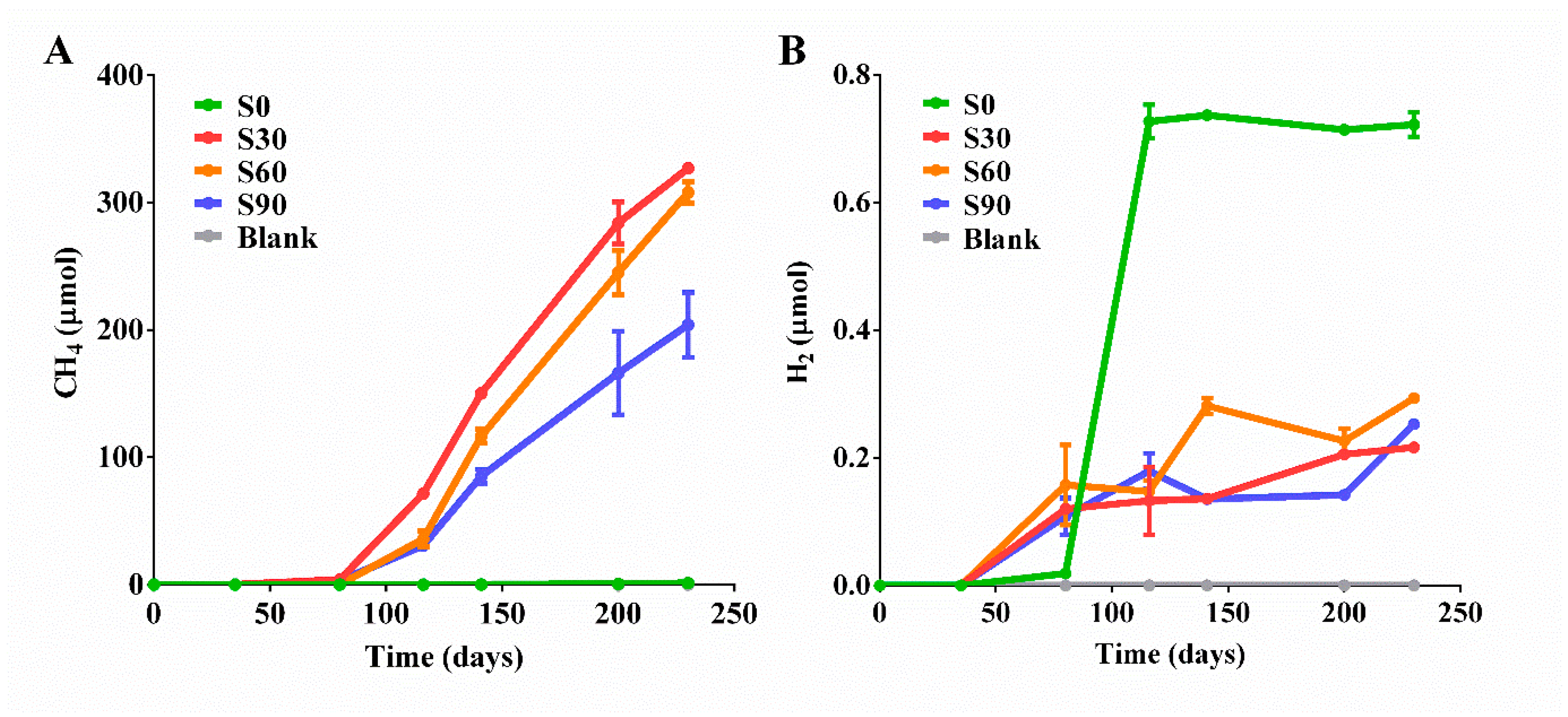


| Samples | Ethanol (d, 0) | Ethanol (d, 230) | Total CH4 | 13CH4 | 12CH4 | CO2 | H2 | Formate | Acetate |
|---|---|---|---|---|---|---|---|---|---|
| S0 | 500 | 297 | 1.25 | 0 | 1.25 | 60.7 | 0.722 | 0.81 | 0.45 |
| S30 | 500 | 123 | 327 | 201 | 126 | 2698 | 0.216 | 3.23 | 6.89 |
| S60 | 500 | 138 | 308 | 211 | 97 | 2515 | 0.293 | 3.42 | 5.77 |
| S90 | 500 | 210 | 204 | 164 | 40 | 2693 | 0.253 | 0.05 | 2.35 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, G.-C.; Zhou, L.; Mbadinga, S.M.; Gu, J.-D.; Mu, B.-Z. Bioconversion Pathway of CO2 in the Presence of Ethanol by Methanogenic Enrichments from Production Water of a High-Temperature Petroleum Reservoir. Energies 2019, 12, 918. https://doi.org/10.3390/en12050918
Yang G-C, Zhou L, Mbadinga SM, Gu J-D, Mu B-Z. Bioconversion Pathway of CO2 in the Presence of Ethanol by Methanogenic Enrichments from Production Water of a High-Temperature Petroleum Reservoir. Energies. 2019; 12(5):918. https://doi.org/10.3390/en12050918
Chicago/Turabian StyleYang, Guang-Chao, Lei Zhou, Serge Maurice Mbadinga, Ji-Dong Gu, and Bo-Zhong Mu. 2019. "Bioconversion Pathway of CO2 in the Presence of Ethanol by Methanogenic Enrichments from Production Water of a High-Temperature Petroleum Reservoir" Energies 12, no. 5: 918. https://doi.org/10.3390/en12050918
APA StyleYang, G.-C., Zhou, L., Mbadinga, S. M., Gu, J.-D., & Mu, B.-Z. (2019). Bioconversion Pathway of CO2 in the Presence of Ethanol by Methanogenic Enrichments from Production Water of a High-Temperature Petroleum Reservoir. Energies, 12(5), 918. https://doi.org/10.3390/en12050918






