Bioengineering and Molecular Biology of Miscanthus
Abstract
:1. Introduction
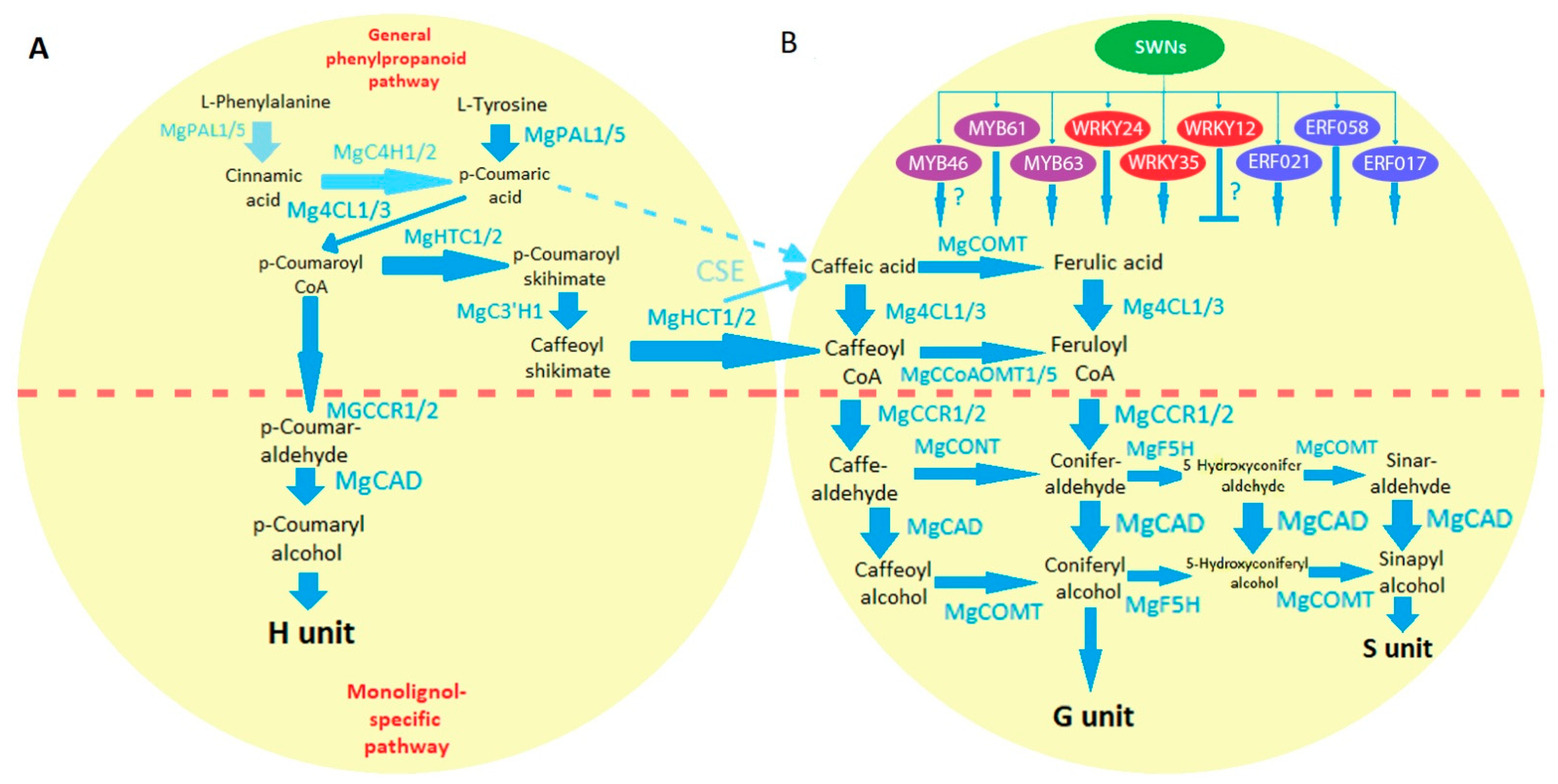
2. The Importance of the Cell Wall in Miscanthus Bioengineering
3. Study of the Miscanthus Genome
4. Cloning of MlARF-GEP, MlKHCP, MlSERK1, MlSERK2 and MlTypA
5. Analysis of Polymorphism of SSR Markers of 5 Genes
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Uvaysova, S.S.; Kaltaeva, S.A. Global fuel and energy complex in the international economy. Innov. Sci. 2015, 4, 3–4. [Google Scholar]
- Wikisource Contributors, «Page:Brundtland Report.djvu/170», Wikisource. Available online: https://en.wikisource.org/w/index.php?title=Page:Brundtland_Report.djvu/170&oldid=7869775 (accessed on 23 June 2022).
- Chomaev, P. Development of timber processing complex of the Russian Federation against the background of universal trends. Univ. Bull. 2017, 10, 47–51. [Google Scholar]
- Babich, O.O.; Krieger, O.V.; Chupakhin, E.G.; Kozlova, O.V. Miscanthus plants processing in fuel, energy, chemical and microbiological industries. Foods Raw Mater. 2019, 7, 411. [Google Scholar] [CrossRef]
- Weijde, T.; Alvim, K.C.; Torres, A.; Vermerris, W.; Dolstra, O.; Visser, R.; Trindade, L. The potential of C4 grasses for cellulosic biofuel production Front. Plant Sci. 2013, 4, 107. [Google Scholar] [CrossRef] [Green Version]
- Heaton, E.; Dohleman, F.; Miguez, F.; Juvik, J.; Lozovaya, V.; Widholm, J.; Zabotina, O.; Mcisaac, G.; Mcisaac, F.; David, M.; et al. Miscanthus: A promising biomass crop. Adv. Bot. Res. 2011, 56, 76. [Google Scholar] [CrossRef]
- Kosolapov, V.; Kostenko, S.; Tyurin, Y.; Shamsutdinova, E.; Piskovskii, Y. Perennial forage grasses—The basis for greening agricultural production. IOP Conf. Ser. Earth Environ. Sci. 2021, 663, 012022. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Strabala, T.J.; Wagner, A. Potential transgenic routes to increase tree biomass. Plant. Sci. 2013, 212, 72–101. [Google Scholar] [CrossRef] [Green Version]
- Mayevskiy, B.Y.; Kurovets, S.S. Hydrocarbons genesis and formation of their deposits as a basis of predicting oil-and-gas presence in deep-sunk horizons of sedimentary basins. EESJ 2016, 8, 122–128. [Google Scholar]
- Pärt, P.; Dorota, E.; Van Dingenen, R.; Larsen, B.; Martini, G. Environment and Human Health—Joint EEA-JRC Report; EUR-OP: London, UK, 2013. [Google Scholar]
- Rakhmankulov, D.L.; Nikolaeva, S.V.; Latypova, F.N.; Vildanov, F.S. On the problem of depletion of world oil reserves. Bashkir. Chem. J. 2008, 15, 5–35. [Google Scholar]
- Christian, D.G.; Riche, A.B.; Yates, N.E. Growth, yield and mineral content of Miscanthus × giganteus grown as a biofuel for 14 successive harvests. Ind. Crops Prod. 2008, 28, 320–327. [Google Scholar] [CrossRef]
- Kukk, M.; Sõber, A. Bud development and shoot morphology in relation to crown location. AoB Plants 2015, 7, plv082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnoult, S.; Brancourt-Hulmel, M. A review on miscanthus biomass production and composition for bioenergy use: Genotypic and environmental variability and implications for breeding. Bioenergy Res. 2015, 8, 502–526. [Google Scholar] [CrossRef] [Green Version]
- Rosen, M.A.; Kishawy, H.A. Sustainable Manufacturing and Design: Concepts, Practices and Needs. Sustainability 2012, 4, 154–174. [Google Scholar] [CrossRef] [Green Version]
- Holechek, J.L.; Geli, H.M.E.; Sawalhah, M.N.; Valdez, R. A Global Assessment: Can Renewable Energy Replace Fossil Fuels by 2050? Sustainability 2022, 14, 4792. [Google Scholar] [CrossRef]
- Nkwachukwu, O.I.; Chima, C.H.; Ikenna, A.O.; Lackson, A. Focus on potential environmental issues on plastic world towards a sustainable plastic recycling in developing countries. Int. J. Ind. Chem. 2013, 4, 34. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Xiao, C.; Xu, S. Starch-Based Completely Biodegradable Polymer Materials. Express Polym. Lett. 2009, 3, 366–375. [Google Scholar] [CrossRef]
- Bhatia, S.; Goli, D. History, scope and development of biotechnology. Introd. Pharm. Biotechnol. 2018, 1, 1–61. [Google Scholar] [CrossRef]
- AgroCounsel.ru. Available online: https://the-farmer.ru/vyraschivanie-miskantusa (accessed on 4 July 2022).
- Devi, A.; Bajar, S.; Kour, H.; Pant, D.; Singh, A. Lignocellulosic Biomass Valorization for Bioethanol Production: A Circular Bioeconomy Approach. Bioenergy Res. 2022. [Google Scholar] [CrossRef]
- Singhvi, M.; Chaudhari, S.; Gokhale, D. Lignocellulose processing: A current challenge. RSC Adv. 2014, 4, 8271–8277. [Google Scholar] [CrossRef]
- Ochoa-Villarreal, M.; Aispuro-Hernández, E.; Vargas-Arispuro, I.; Martínez-Téllez, M.Á. Plant Cell Wall Polymers: Function, Structure and Biological Activity of Their Derivatives. In Polymerization; IntechOpen: London, UK, 2012. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Li, L.; Xu, F. Chemical Characteristics of Wood Cell Wall with an Emphasis on Ultrastructure: A Mini-Review. Forests 2022, 13, 439. [Google Scholar] [CrossRef]
- Sainz-Polo, M.A.; Valenzuela, S.V.; González, B.; Pastor, F.I.; Sanz-Aparicio, J. Structural analysis of glucuronoxylan-specific Xyn30D and its attached CBM35 domain gives insights into the role of modularity in specificity. J. Biol. Chem. 2014, 289, 31088–31101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yapo, B. Pectic substances: From simple pectic polysaccharides to complex pectins—A new hypothetical model. Carbohyd. Polym. 2011, 86, 373–385. [Google Scholar] [CrossRef]
- Sluiter, A.; Hames, B.; Ruiz, R.; Scarlata, C.; Sluiter, J.; Templeton, D.; Crocker, D. Determination of Structural Carbohydrates and Lignin in Biomass—NREL/TP-510-42618. Lab. Anal. Proced. 2008, 1617, 1–16. [Google Scholar]
- Kaack, K.; Schwarz, K.-U.; Brander, P.E. Variation in morphology, anatomy and chemistry of stems of Miscanthus genotypes differing in mechanical properties. Ind. Crops Prod. 2003, 17, 131–142. [Google Scholar] [CrossRef]
- Gismatulina, Y.A.; Budaeva, V.V. Chemical Composition of Russian Miscanthus and the Quality of Cellulose Obtained Therefrom. Chem. Sustain. Dev. 2013, 21, 503–508. [Google Scholar]
- Klímek, P.; Wimmer, R.; Meinlschmidt, P.; Kúdela, J. Utilizing Miscanthus stalks as raw material for particleboards. Ind. Crops Prod. 2018, 111, 270–276. [Google Scholar] [CrossRef]
- Boneca, I.G.; Huang, Z.-H.; Gage, D.A.; Tomasz, A. Characterization of Staphylococcus aureus Cell Wall Glycan Strands, Evidence for a New β-N-Acetylglucosaminidase Activity. J. Biol. Chem. 2000, 275, 9910–9918. [Google Scholar] [CrossRef] [Green Version]
- Da Costa, R.M.; Pattathil, S.; Avci, U.; Lee, S.J.; Hazen, S.P.; Winters, A.; Hahn, M.G.; Bosch, M. A cell wall reference profile for Miscanthus bioenergy crops highlights compositional and structural variations associated with development and organ origin. New Phytol. 2017, 213, 1710–1725. [Google Scholar] [CrossRef]
- Kumar, A.; Choudhary, A.K.; Pooniya, V.; Suri, V.K.; Singh, U. Soil Factors Associated with Micronutrient Acquisition in Crops- Biofortification Perspective. In Biofortification of Food Crops; Singh, U., Praharaj, C., Singh, S., Singh, N., Eds.; Springer: New Delhi, India, 2016. [Google Scholar] [CrossRef]
- Clark, L.V.; Dwiyanti, M.S.; Anzoua, K.G.; Brummer, J.E.; Ghimire, B.K.; Głowacka, K.; Hall, M.; Heo, K.; Jin, X.; Lipka, A.E.; et al. Genome-wide association and genomic prediction for biomass yield in a genetically diverse Miscanthus sinensis germplasm panel phenotyped at five locations in Asia and North America. GCB Bioenergy 2019, 8, 585. [Google Scholar] [CrossRef] [Green Version]
- Mitros, T.; Session, A.M.; James, B.T.; Wu, G.A.; Belaffif, M.B.; Clark, L.V.; Shu, S.; Dong, H.; Barling, A.; Holmes, J.R.; et al. Genome biology of the paleotetraploid perennial biomass crop Miscanthus. Nat. Commun. 2020, 11, 5442. [Google Scholar] [CrossRef]
- Van der Weijde, T.; Kamei, C.L.A.; Severing, E.I.; Torres, A.F.; Gomez, L.D.; Dolstra, O.; Maliepaard, C.A.; McQueen-Mason, S.J.; Visser, R.G.F.; Trindade, L.M. Genetic complexity of miscanthus cell wall composition and biomass quality for biofuels. BMC Genom. 2017, 18, 406. [Google Scholar] [CrossRef] [PubMed]
- Moreno, A.; Alvira, P.; Ibarra, D.; Tomas-Pejo, E. Production of Ethanol from Lignocellulosic Biomass. In Production of Platform Chemicals from Sustainable Resources; Fang, Z., Smith, R.L., Qui, X., Eds.; Biofuels and Biorefineries Chapter: 12; Springer: Singapore, 2017; Volume 7, pp. 375–410. [Google Scholar] [CrossRef]
- Ten, E.; Vermerris, W. Functionalized Polymers from Lignocellulosic Biomass: State of the Art. Polymers 2013, 5, 600–642. [Google Scholar] [CrossRef] [Green Version]
- Tao, W.; Xin, L. Overcome saccharification barrier: Advances in hydrolysis technology. In Woodhead Publishing Series in Energy, Advances in 2nd Generation of Bioethanol Production; Xin, L., Ed.; Woodhead Publishing: Thorston, UK, 2021; pp. 137–159. [Google Scholar] [CrossRef]
- Zeng, X.; Sheng, J.; Zhu, F.; Wei, T.; Zhao, L.; Hu, X.; Zheng, X.; Zhou, F.; Hu, Z.; Diao, Y.; et al. Genetic, transcriptional, and regulatory landscape of monolignol biosynthesis pathway in Miscanthus × giganteus. Biotechnol. Biofuels 2020, 13, 179. [Google Scholar] [CrossRef]
- Anderson, N.A.; Tobimatsu, Y.; Ciesielski, P.N.; Ximenes, E.; Ralph, J.; Donohoe, B.S.; Ladisch, M.; Chapple, C. Manipulation of Guaiacyl and Syringyl Monomer Biosynthesis in an Arabidopsis Cinnamyl Alcohol Dehydrogenase Mutant Results in Atypical Lignin Biosynthesis and Modified Cell Wall Structure. Plant Cell. 2015, 27, 2195–2209. [Google Scholar] [CrossRef] [PubMed]
- Sundin, L.; Vanholme, R.; Geerinck, J.; Goeminne, G.; Höfer, R.; Kim, H.; Ralph, J.; Boerjan, W. Mutation of the inducible Arabidopsis thaliana cytochrome P450 REDUCTASE2 alters lignin composition and improves saccharification. Plant Physiol. 2014, 166, 1956–1971. [Google Scholar] [CrossRef] [Green Version]
- Barros, J.; Escamilla-Trevino, L.; Song, L.; Rao, X.; Serrani-Yarce, J.C.; Palacios, M.D.; Engle, N.; Choudhury, F.K.; Tschaplinski, T.J.; Venables, B.J.; et al. 4-Coumarate 3-hydroxylase in the lignin biosynthesis pathway is a cytosolic ascorbate peroxidase. Nat. Commun. 2019, 10, 1994. [Google Scholar] [CrossRef] [Green Version]
- Zubieta, C.; Kota, P.; Ferrer, J.L.; Dixon, R.A.; Noel, J.P. Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell. 2002, 14, 1265–1277. [Google Scholar] [CrossRef] [Green Version]
- Hill, A.M.; Staunton, J. Type I Modular PKS. Chem. Biol. 2010, 1, 385–452. [Google Scholar] [CrossRef]
- Lee, Y.; Chen, F.; Gallego-Giraldo, L.; Dixon, R.A.; Voit, E.O. Integrative Analysis of Transgenic Alfalfa (Medicago sativa L.) Suggests New Metabolic Control Mechanisms for Monolignol Biosynthesis. PLoS Comput. Biol. 2011, 7, e1002047. [Google Scholar] [CrossRef]
- Hyun, M.W.; Yun, Y.H.; Kim, J.Y.; Kim, S.H. Fungal and Plant Phenylalanine Ammonia-lyase. Mycobiology 2011, 39, 257–265. [Google Scholar] [CrossRef] [Green Version]
- Decena, M.A.; Gálvez-Rojas, S.; Agostini, F.; Sancho, R.; Contreras-Moreira, B.; Des Marais, D.L.; Hernandez, P.; Catalán, P. Comparative Genomics, Evolution, and Drought-Induced Expression of Dehydrin Genes in Model Brachypodium Grasses. Plants 2021, 10, 2664. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zheng, W.; Zhao, H.; Deng, M. Statistical Analysis Reveals Co-Expression Patterns of Many Pairs of Genes in Yeast Are Jointly Regulated by Interacting Loci. PLoS Genet. 2013, 9, e1003414. [Google Scholar] [CrossRef]
- Wang, G.-Z.; Du, K.; Hu, S.-Q.; Chen, S.-Y.; Jia, X.-B.; Cai, M.-C.; Shi, Y.; Wang, J.; Lai, S.-J. Genome-wide identification and characterization of long non-coding RNAs during postnatal development of rabbit adipose tissue. Lipids Health Dis. 2018, 17, 271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chupakhin, E.; Babich, O.; Sukhikh, S.; Ivanova, S.; Budenkova, E.; Kalashnikova, O.; Kriger, O. Methods of Increasing Miscanthus Biomass Yield for Biofuel Production. Energies 2021, 14, 8368. [Google Scholar] [CrossRef]
- Clark, L.V.; Stewart, J.R.; Nishiwaki, A.; Toma, Y.; Kjeldsen, J.B.; Jørgensen, U.; Zhao, H.; Peng, J.; Yoo, J.H.; Heo, K.; et al. Genetic structure of Miscanthus sinensis and Miscanthus sacchariflorus in Japan indicates a gradient of bidirectional but asymmetric introgression. J. Exp. Bot. 2015, 66, 4213–4225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hodkinson, T.; Chase, M.; Lledó, M.; Salamin, N.; Renvoize, S. Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences from ITS nuclear ribosomal DNA and plastid trnL intron and trnL-F intergenic spacers. J. Plant. Res. 2002, 115, 381–392. [Google Scholar] [CrossRef]
- Tamura, K.-i.; Uwatoko, N.; Yamashita, H.; Fujimori, M.; Akiyama, Y.; Shoji, A.; Sanada, Y.; Okumura, K.; Gau, M. Discovery of Natural Interspecific Hybrids Between Miscanthus Sacchariflorus and Miscanthus Sinensis in Southern Japan: Morphological Characterization, Genetic Structure, and Origin. Bioenergy Res. 2016, 9, 315–325. [Google Scholar] [CrossRef]
- Kovalchuk, N.; Roik, M. Miscanthus: Genetic diversity and a method of ploidy variability identification using fluorescent cytophotometry. Agric. Sci. Pract. 2017, 4, 19–27. [Google Scholar] [CrossRef]
- Maughan, M.; Bollero, G.; Lee, D.; Darmody, R.; Bonos, S.; Cortese, L.; Murphy, J.; Gaussoin, R.; Sousek, M.; Williams, D.; et al. Miscanthus × giganteus productivity: The effects of management in different environments. GCB Bioenergy 2012, 4, 253–265. [Google Scholar] [CrossRef]
- Nishiwaki, A.; Mizuguti, A.; Kuwabara, S.; Toma, Y.; Ishigaki, G.; Miyashita, T.; Yamada, T.; Matuura, H.; Yamaguchi, S.; Rayburn, A.; et al. Discovery of natural Miscanthus (Poaceae) triploid plants in sympatric populations of Miscanthus sacchariflorus and Miscanthus sinensis in southern Japan. Am. J. Bot. 2011, 98, 154–159. [Google Scholar] [CrossRef]
- St Charles, J.; Hamilton, M.L.; Petes, T.D. Meiotic chromosome segregation in triploid strains of Saccharomyces cerevisiae. Genetics 2010, 186, 537–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kar, S.; Zhang, N.; Nakashima, T.; Villanueva-Morales, A.; Stewart, J.; Sacks, E.; Terajima, Y.; Yamada, T. Saccharum × Miscanthus intergeneric hybrids (miscanes) exhibit greater chilling tolerance of C4 photosynthesis and postchilling recovery than sugarcane (Saccharum spp. hybrids). GCB Bioenergy 2019, 11, 1318–1333. [Google Scholar] [CrossRef] [Green Version]
- Kołodziej, B.; Antonkiewicz, J.; Sugier, D. Miscanthus × giganteus as a biomass feedstock grown on municipal sewage sludge. Ind. Crops Prod. 2016, 81, 72–82. [Google Scholar] [CrossRef]
- Jolliffe, I.T.; Cadima, J. Principal component analysis: A review and recent developments. Philos. Trans. R. Soc. A 2016, 374, 20150202. [Google Scholar] [CrossRef]
- Zhao, L.; Zeng, X.; Hu, X.; Sheng, J.; Zhu, F.; Zhong, L.; Zhou, F.; Jin, S.; Hu, Z.; Diao, Y. Molecular cloning and characterization of five genes from embryogenic callus in Miscanthus lutarioriparius. Acta Physiol. Plant. 2020, 42, 89. [Google Scholar] [CrossRef]
- Sun, L.; Wu, Y.; Su, S.; Liu, H.; Yang, G.; Li, S.; Shan, X.; Yuan, Y. Differential gene expression during somatic embryogenesis in the maize (Zea mays L.) inbred line H99. PCTOC 2011, 109, 271–286. [Google Scholar] [CrossRef]
- Xia, Y.; Xu, J.; Duan, J.; Liu, Q.; Huang, H.; Yi, Z.; Chen, Z. Transgenic Miscanthus lutarioriparius that co-expresses the Cry 2Aa# and Bar genes. Can. J. Plant. Sci. 2019, 99, 841–851. [Google Scholar] [CrossRef]
- Woldesemayat, A.A.; Van Heusden, P.; Ndimba, B.K.; Christoffels, A. An integrated and comparative approach towards identification, characterization and functional annotation of candidate genes for drought tolerance in sorghum (Sorghum bicolor (L.) Moench). BMC Genet. 2017, 18, 119. [Google Scholar] [CrossRef] [Green Version]
- Park, C.-H.; Kim, Y.-G.; Kim, K.-H.; Alam, I.; Lee, H.-J.; Sharmin, S.; Lee, K.-W.; Lee, B.-H. Effect of Plant Growth Regulators on Callus Induction and Plant Regeneration from Mature Seed Culture of Miscanthus sinensis. J. Korean Soc. Grassl. Forage Sci. 2009, 29, 291–298. [Google Scholar] [CrossRef]
- Glowacka, K.; Jezowski, S.; Kaczmarek, Z. The effects of genotype, inflorescence developmental stage and induction medium on callus induction and plant regeneration in two Miscanthus species. PCTOC 2010, 102, 79–86. [Google Scholar] [CrossRef]
- Zhang, Q.; Sun, Y.; Hu, H.; Chen, B.; Hong, C.; Guo, H.; Pan, Y.; Zheng, B. Micropropagation and plant regeneration from embryogenic callus of Miscanthus sinensis. In Vitro Cell. Dev. Biol.-Plant 2012, 48, 50–57. [Google Scholar] [CrossRef]
- Rambaud, C.; Arnoult, S.; Bluteau, A.; Mansard, M.C.; Blassiau, C.; Brancourt-Hulmel, M. Shoot organogenesis in three Miscanthus species and evaluation for genetic uniformity using AFLP analysis. PCTOC 2013, 113, 437–448. [Google Scholar] [CrossRef] [Green Version]
- Salehi, H.; Khosh-Khui, M. Effects of genotype and plant growth regulator on callus induction and plant regeneration in four important turegrass genera: A comparative study. In Vitro Cell. Dev. Biol.-Plant 2005, 41, 157–161. [Google Scholar] [CrossRef]
- Frank, S.; Upender, S.; Hansen, S.H.; Casanova, J.E. ARNO is a guanine nucleotide exchange factor for ADP-ribosylation factor 6. J. Biol. Chem. 1998, 273, 23–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blinstrubienė, A.; Jančauskienė, I.; Burbulis, N. In Vitro Regeneration of Miscanthus × giganteus through Indirect Organogenesis: Effect of Explant Type and Growth Regulators. Plants 2021, 10, 2799. [Google Scholar] [CrossRef]
- Liu, P.; Myo, T.; Ma, W.; Lan, D.; Qi, T.; Guo, J.; Song, P.; Guo, J.; Kang, Z. TaTypA, a Ribosome-Binding GTPase Protein, Positively Regulates Wheat Resistance to the Stripe Rust Fungus. Front. Plant. Sci. 2016, 7, 873. [Google Scholar] [CrossRef] [Green Version]
- Dou, X.Y.; Yang, K.Z.; Ma, Z.X.; Chen, L.Q.; Zhang, X.Q.; Bai, J.R.; Ye, D. AtTMEM18 plays important roles in pollen tube and vegetative growth in Arabidopsis. J. Integr. Plant. Biol. 2016, 58, 679–692. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.; Muszynski, M.G.; Danilevskaya, O.N. The FT-like ZCN8 Gene Functions as a Floral Activator and Is Involved in Photoperiod Sensitivity in Maize. Plant. Cell. 2011, 23, 942–960. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Zhang, G.; Muhammad, A.; Samad, R.A.; Wang, Y.; Walton, J.D.; He, Y.; Peng, L.; Wang, L. Genetic loci simultaneously controlling lignin monomers and biomass digestibility of rice straw. Sci. Rep. 2018, 8, 3636. [Google Scholar] [CrossRef]
- Heaton, E.; Long, S.; Voigt, T.; Jones, M.; Clifton-Brown, J. Miscanthus for Renewable Energy Generation: European Union Experience and Projections for Illinois. Mitig. Adapt. Strateg. Glob. Chang. 2004, 9, 433–451. [Google Scholar] [CrossRef]
- Guimarães, C.T.; Sills, G.R.; Sobral, B.W. Comparative mapping of Andropogoneae: Saccharum L. (sugarcane) and its relation to sorghum and maize. Proc. Natl. Acad. Sci. USA 1997, 94, 14261–14266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, H.; Clark, L.; Jin, X.; Anzoua, K.; Larisa, B.; Chebukin, P.; Dzyubenko, E.A.; Dzyubenko, N.; Ghimire, B.; Heo, K.; et al. Managing flowering time in Miscanthus and sugarcane to facilitate intra- and intergeneric crosses. PLoS ONE 2021, 16, e0240390. [Google Scholar] [CrossRef] [PubMed]
| Type of Miscanthus | Mass Fraction of the Component, % | |||||
|---|---|---|---|---|---|---|
| Adipose Fraction | Ash | Lignin | Pentosans | Cellulose * | Extractive Substances | |
| M. × giganteus | 4.37 ± 0.16 | 5.12 ± 0.19 | 22.14 ± 0.66 | 23.62 ± 0.69 | 42.00 ± 1.26 | 5.7 ± 0.1 |
| M. sinensis | 4.81 ± 0.14 | 6.05 ± 0.18 | 23.79 ± 0.67 | 23.17 ± 0.69 | 43.63 ± 1.30 | 2.8 ± 0.1 |
| M. sacchariflonis | 5.05 ± 0.15 | 6.20 ± 0.18 | 22.11 ± 0.66 | 25.47 ± 0.69 | 41.98 ± 1.26 | 4.1 ± 0.1 |
| Type of Miscanthus | Ploidy | Productivity | Amount of Biomass: Raw/Dry, t/ha | Sources |
|---|---|---|---|---|
| M. × giganteus | triploid | highly productive | 10.71/3.53 | [56] |
| M. sacchariflorus | tetraploid | highly productive | 9.72/2.97 | [52] |
| M. sinensis | diploid | productive | 7.76/2.36 | [52] |
| Month | C/N Ratio | ||
|---|---|---|---|
| 1 | 2 | 3 | |
| August | 95.2 | 97.5 | 99.2 |
| September | 103.1 | 109.5 | 120.3 |
| October | 108.7 | 121.8 | 160.3 |
| November | 113.6 | 148.9 | 204.1 |
| December | 129.8 | 188.4 | 264.0 |
| January | 135.4 | 201.7 | 300.5 |
| February | 140.1 | 212.0 | 322.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chupakhin, E.; Babich, O.; Sukhikh, S.; Ivanova, S.; Budenkova, E.; Kalashnikova, O.; Prosekov, A.; Kriger, O.; Dolganyuk, V. Bioengineering and Molecular Biology of Miscanthus. Energies 2022, 15, 4941. https://doi.org/10.3390/en15144941
Chupakhin E, Babich O, Sukhikh S, Ivanova S, Budenkova E, Kalashnikova O, Prosekov A, Kriger O, Dolganyuk V. Bioengineering and Molecular Biology of Miscanthus. Energies. 2022; 15(14):4941. https://doi.org/10.3390/en15144941
Chicago/Turabian StyleChupakhin, Evgeny, Olga Babich, Stanislav Sukhikh, Svetlana Ivanova, Ekaterina Budenkova, Olga Kalashnikova, Alexander Prosekov, Olga Kriger, and Vyacheslav Dolganyuk. 2022. "Bioengineering and Molecular Biology of Miscanthus" Energies 15, no. 14: 4941. https://doi.org/10.3390/en15144941









