Low and High Field Magnetic Resonance for in Vivo Analysis of Seeds
Abstract
:1. Introduction
2. Methods
2.1. Hardware
2.2. Plant Seed Material
2.3. Application of MQ60
2.4. Lipid Mapping of Mature Seeds Using 1H MRS-Imaging
2.5. Localized 1H NMR and 1H Spectroscopic Imaging on Fresh Harvested Seed
2.6. Double Resonant 13C / 1H NMR for Tracing 13C Metabolites in Developing Seeds
3. Results and Discussion
3.1. Low Field NMR Enables Accurate and Fast Quantification of Seed Lipids in a Wide Range of Species

3.2. High Field MRI is a Versatile Tool for Exploring Grain Lipid Composition


3.3. Localized 1H NMR and 1H Spectroscopic Imaging Allows Assessment of Metabolite Distribution within the Living Seed
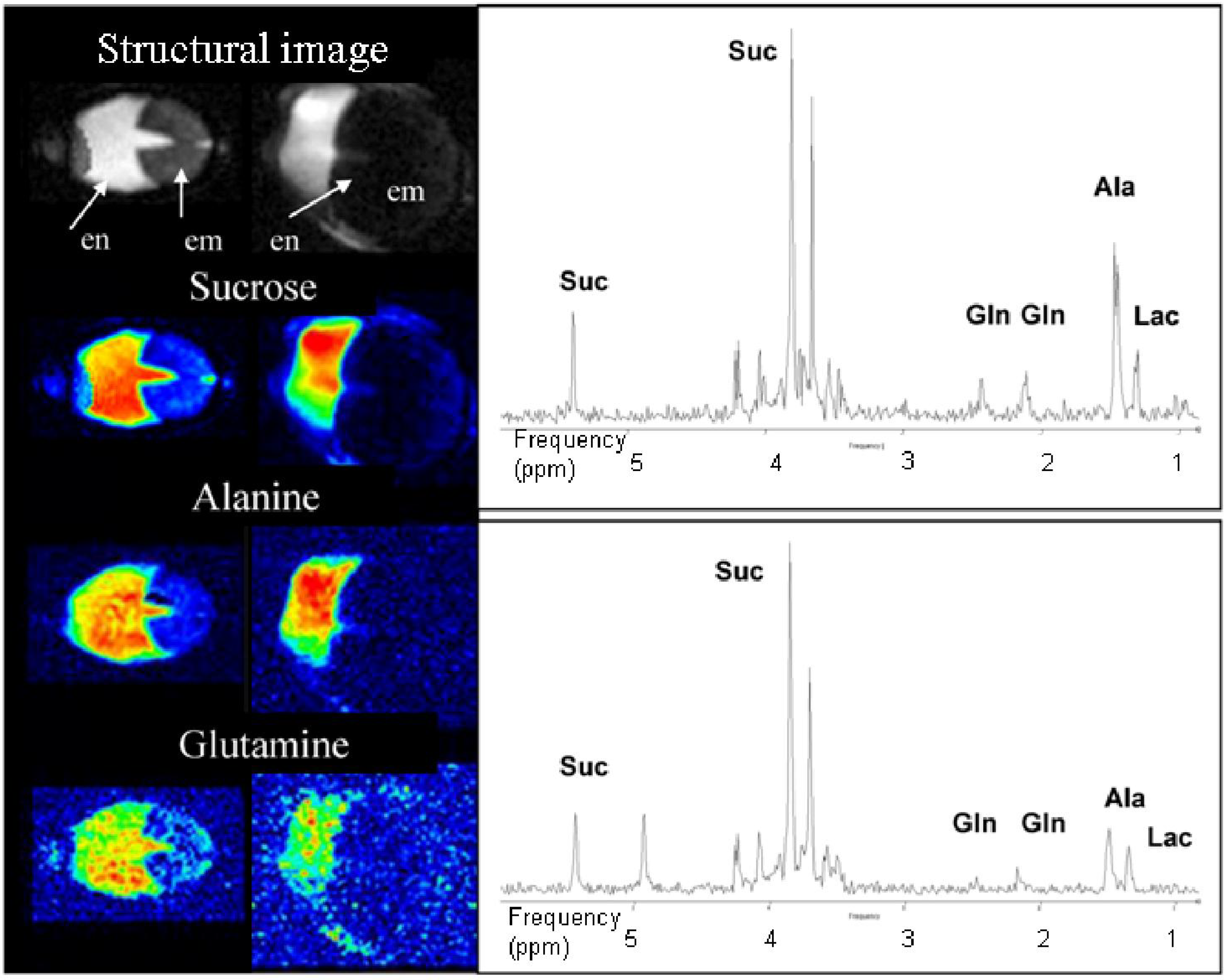

3.4. Double Resonant 1H /13C NMR Allows the Tracing of 13C Labelled Metabolites within the Seed

4. Conclusions
Acknowledgments
References
- Krygsman, P.H.; Barrett, A.E. Simple methods for measuring total oil content by benchtop NMR. Oil Extraction and Analisis 2004, 152–165. [Google Scholar]
- Todt, H.; Guthausen, G.; Burk, W.; Schmalbein, D.; Kamlowski, A. Water/moisture and fat analysis by time-domain NMR. Food Chem. 2005, 96, 436–440. [Google Scholar] [CrossRef]
- Ghosh, S.; Tombokan, X. TD-NMR technology: A noninvasive tool for high-throughput QC and rapid cereal product improvement. Cereal Foods World 2009, 54, 152–157. [Google Scholar]
- Van Duynhoven, J.; Voda, A.; Witek, M.; van As, H. Time-domain Nmr applied to food products. Ann. Rep. NMR Spectrsp. 2010, 69, 145–197. [Google Scholar]
- U.S. Department of Agriculture GIPSA. GIPSA Procedure for Determining the Oil Content of Pre-Dried Sunflower Seed Using the Bruker Minispec 7.5 Pulsed NMR Instrument; Certificate No. FGIS 00-101; U.S. Department of Agriculture: Washington, DC, USA, 1999. [Google Scholar]
- International Standard Organization (ISO) 10565. Oilseeds-Simultaneous Determination of Oil and Moisture Contents-Method Using Pulsed Nuclear Magnetic Resonance Spectrometry; International Organization for Standardization: Geneva, Switzerland, 1999. [Google Scholar]
- Vaughan, J.T.; Hetherington, H.P.; Out, J.O.; Pan, J.W.; Pohost, G.M. High frequency volume coils for clinical NMR imaging and spectroscopy. Magn. Reson. Med. 1994, 32, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Köckenberger, W.; DePanfilis, C.; Santoro, D.; Dahiva, P.; Rawsthorne, S. High resolution NMR microscopy of plants and fungi. J. Microsc. 2004, 214, 182–189. [Google Scholar] [CrossRef]
- Judith, R.; Rathjen, E.; Strounina, V.; Mares, D.J. Water movement into dormant and non-dormant wheat (Triticum aestivum L.) grains. J. Exp. Bot. 2009, 60, 1619–1631. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, M.; Kikuchi, K.; Isobe, S.; Ishida, N.; Naito, S.; Kano, H. Role of seed coat in imbibing soybean seeds observed by micro-magnetic resonance imaging. Ann Bot. 2008, 102, 343–352. [Google Scholar] [CrossRef] [PubMed]
- Lakshinarayana, M.R.; Syamasudar, J.; Gowda, G.; Khetrapal, C.L. Spatial distribution of oil in groundnut and sunflower seeds by nuclear magnetic resonance imaging. J. Biosci. 1992, 17, 87–93. [Google Scholar]
- Ischida, N.; Koizumi, M.; Kano, H. The NMR microscope: A unique and promising tool for plant science. Ann. Bot. 2000, 86, 259–278. [Google Scholar] [CrossRef]
- Baranska, M.; Schulz, H.; Reitzenstein, S.; Uhlemann, U.; Strehle, M.A.; Kruger, H.; Quilitzsch, R.; Foley, W.; Popp, J. Vibrational spectroscopic studies to acquire a quality control method of Eucalyptus essential oils. Biopolymers 2005, 78, 237–248. [Google Scholar] [CrossRef] [PubMed]
- Knothe, G.; Kenar, J.A. Determination of the fatty acid profile by 1H-NMR spectroscopy. Eur. J. Lipid Sci. Technol. 2004, 106, 88–96. [Google Scholar] [CrossRef]
- Borisjuk, L.; Nguyen, T.H.; Neuberger, T.; Rutten, T.; Tschiersch, H.; Claus, B.; Feussner, I.; Webb, A.G.; Jakob, P.; Weber, H.; Wobus, U.; Rolletschek, H. Gradients of lipid storage, photosynthesis and plastid differentiation in developing soybean seeds. New Phytol. 2005, 167, 761–776. [Google Scholar] [CrossRef] [PubMed]
- Tse, T.Y.; Spanswick, R.M.; Jelinski, L.W. Quantitative evaluation of NMR and MRI methods to measure sucrose concentrations in plants. Protoplasma 1996, 194, 54–62. [Google Scholar] [CrossRef]
- Melkus, G.; Rolletschek, H.; Radchuk, R.; Fuchs, J.; Rutten, T.; Wobus, U.; Altmann, T.; Jakob, P.; Borisjuk, L. The metabolic role of legume endosperm: A non-invasive imaging study. Plant Physiol. 2009, 151, 1139–1154. [Google Scholar] [CrossRef] [PubMed]
- Kalusche, B.; Verscht, J.; Gebauer, G.; Komor, E.; Haase, A. Sucrose unloading in the hypocotyl of the Ricinus communis L. seedling measured by 13C-nuclear magnetic resonance spectroscopy in vivo. Planta 1999, 208, 358–364. [Google Scholar] [CrossRef]
- Ratcliffe, R.G.; Roscher, A.; Shachar-Hill, Y. Plant NMR spectroscopy. Prog. Nucl. Magn. Reson. Sp. 2001, 39, 267–300. [Google Scholar] [CrossRef]
- Köckenberger, W. Functional imaging of plants by magnetic resonance experiments. Trends Plant Sci. 2001, 6, 286–292. [Google Scholar] [CrossRef] [PubMed]
- Bardet, M.; Foray, M.F.; Guillermo, A. High-resolution solid-state NMR as an analytical tool to study plant seeds. Modern Magn. Reson. 2006, 7, 1777–1781. [Google Scholar]
- Terskikh, V.V.; Feurtado, J.A.; Ren, C.; Abrams, S.R.; Kermode, A.R. Water uptake and oil distribution during imbibition of seeds of western white pine (Pinus monticola Dougl. ex D. Don) monitored in vivo using magnetic resonance imaging. Planta 2005, 221, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Annarao, S.; Sidhu, O.P.; Roy, R.; Tuli, R.; Khetrapal, C.L. Lipid profiling of developing Jatropha curcas L. seeds using 1H NMR spectroscopy. Bioresource Technol. 2008, 99, 9032–9035. [Google Scholar] [CrossRef]
- Neuberger, T.; Sreenivasulu, N.; Rokitta, M.; Rolletschek, H.; Göbel, C.; Rutten, T.; Radchuk, V.; Feussner, I.; Wobus, U.; Jakob, P.; Borisjuk, L. Quantitative imaging of oil storage in developing crop seeds. Plant Biotechnol. J. 2008, 6, 31–45. [Google Scholar] [PubMed]
- Minchin, P.E.H.; Thorpe, M.R. Using the short-lived isotope 11C in mechanistic studies of photosynthate transport. Funct. Plant Biol. 2003, 30, 831–841. [Google Scholar] [CrossRef]
- Jahnke, S.; Menzel, M.I.; van Dusschoten, D.; Roeb, G.W.; Bühler, J.; Minwuyelet, S.; Blümler, P.; Temperton, V.M.; Hombach, T.; Streun, M.; et al. Combined MRI-PET dissects dynamic changes in plant structures and functions. Plant J. 2009, 59, 634–644. [Google Scholar] [CrossRef]
- Melkus, G.; Rolletschek, H.; Fuchs, J.; Radchuk, V.; Grafahrend-Belau, E.; Sreenivasulu, N.; Rutten, T.; Weier, D.; Heinzel, N.; Schreiber, F.; et al. Dynamic 13C/1H NMR imaging uncovers sugar allocation in the living seed. Plant Biotechnol. J. 2011. [Google Scholar] [CrossRef]
- Nagel, M.; Vogel, H.; Landjeva, S.; Buck-Sorlin, G.; Lohwasser, U.; Scholz, U.; Börner, A. Seed conservation in ex-situ genebanks-genetic studies on longevity in barley. Euphytica 2009, 170, 1–10. [Google Scholar] [CrossRef]
- Neuberger, T.; Rolletschek, H.; Webb, A.; Borisjuk, L. Non-invasive mapping of lipids in plant tissue using magnetic resonance imaging. Methods Mol. Biol. 2009, 579, 485–496. [Google Scholar]
- Seal, C.E.; Kranner, I.; Pritchard, H.W. Quantification of seed oil from species with varying oil content using supercritical fluid extraction. Phytochem. Anal. 2008, 19, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.R.; Kincaid, B.M.; Ugurbil, K. NMR chemical shift imaging in three dimensions. Proc. Natl. Acad. Sci. USA 1982, 79, 3523–3526. [Google Scholar] [CrossRef] [PubMed]
- Tkác, I.; Starcuk, Z.; Choi, I.Y.; Gruetter, R. In vivo 1H NMR spectroscopy of rat brain at 1 ms echo time. Magn. Reson. Med. 1999, 41, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Soher, B.J.; van Zijl, P.C.; Duyn, J.H.; Barker, P.B. Quantitative proton MR spectroscopic imaging of the human brain. Magn. Reson. Med. 1996, 35, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Thomas, M.A.; Yue, K.; Binesh, N. Localized two-dimensional shift correlated MR spectroscopy of human brain. Magn. Reson. Med. 2001, 46, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Hurd, R.E.; John, B.K. Gradient-enhanced proton-detected heteronuclear multiple-quantum coherence spectroscopy. J. Magn. Resonance 1991, 91, 648–653. [Google Scholar]
- Novotny, E.J., Jr.; Ogino, T.; Rothman, D.L.; Petroff, O.A.; Prichard, J.W.; Shulman, R.G. Direct carbon versus proton heteronuclear editing of 2-13C ethanol in rabbit brain in vivo: A sensitivity comparison. Magn. Reson. Med. 1990, 16, 431–43. [Google Scholar] [CrossRef] [PubMed]
- Rance, M.; Byrd, R.A. Obtaining high-fidelity spin-1/2 powder spectra in anisotropic media: Phase-cycled Hahn Echo spectroscopy. J. Magn. Resonance 1983, 52, 221–240. [Google Scholar]
- Davis, D.G. Elimination of baseline distortions and minimization of artefacts from phased 2D NMR spectra. J. Magn. Resonance 1989, 81, 603–607. [Google Scholar]
- McDonald, M.B. Seed deterioration: Physiology, repair and assessment. Seed Sci. Technol. 1999, 27, 177–237. [Google Scholar]
- Terskikh, V.V.; Zeng, Y.; Feurtado, J.A.; Giblin, M.; Abrams, S.R.; Kermode, A.R. Deterioration of western redcedar (Thuja plicata Donn ex D. Don) seeds: Protein oxidation and in vivo NMR monitoring of storage oils. J. Exp. Bot. 2008, 59, 765–777. [Google Scholar] [CrossRef] [PubMed]
- Walters, C.; Wheeler, L.M.; Grotenhuis, J.M. Longevity of seeds stored in a genebank: Species characteristics. Seed Sci. Res. 2005, 15, 1–20. [Google Scholar] [CrossRef]
- Chen-Miller, J.; Schopf, W.J.; Harbottle, G.; Cao, R.; Ouyang, S.; Zhou, K.; Southon, J.R.; Guo-hai, L. Long-living lotus: Germination and soil γ-irradiation of centuries-old fruits, and cultivation, growth, and phenotypic abnormalities of offspring. Am. J. Bot. 2002, 89, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Alonso-Blanco, C.; Blankestijn-de Vries, H.; Hanhart, C.J.; Koornneef, M. Natural allelic variation at seed size loci in relation to other life history traits of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1999, 96, 4710–4717. [Google Scholar] [CrossRef] [PubMed]
- Melkus, G.; Mörchel, P.; Behr, V.C.; Kotas, M.; Flentje, M.; Jakob, P.M. Short echo spectroscopic imaging combined with lactate editing in a single scan. NMR Biomed. 2008, 21, 1076–1086. [Google Scholar] [CrossRef] [PubMed]
- Weiss, K.; Melkus, G.; Jakob, P.M.; Faber, C. Quantitative in vivo 1H spectroscopic imaging of metabolites in the early postnatal mouse brain at 17.6 T. Magma 2009, 22, 62–72. [Google Scholar] [CrossRef]
- Fisher, D.B.; Gifford, R.M. Accumulation and conversion of sugars by developing wheat grains: VI. Gradients along the transport pathway from the peduncle to the endosperm cavity during grain filling. Plant Physiol. 1986, 82, 1024–1030. [Google Scholar] [CrossRef] [PubMed]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Borisjuk, L.; Rolletschek, H.; Fuchs, J.; Melkus, G.; Neuberger, T. Low and High Field Magnetic Resonance for in Vivo Analysis of Seeds. Materials 2011, 4, 1426-1439. https://doi.org/10.3390/ma4081426
Borisjuk L, Rolletschek H, Fuchs J, Melkus G, Neuberger T. Low and High Field Magnetic Resonance for in Vivo Analysis of Seeds. Materials. 2011; 4(8):1426-1439. https://doi.org/10.3390/ma4081426
Chicago/Turabian StyleBorisjuk, Ljudmilla, Hardy Rolletschek, Johannes Fuchs, Gerd Melkus, and Thomas Neuberger. 2011. "Low and High Field Magnetic Resonance for in Vivo Analysis of Seeds" Materials 4, no. 8: 1426-1439. https://doi.org/10.3390/ma4081426



