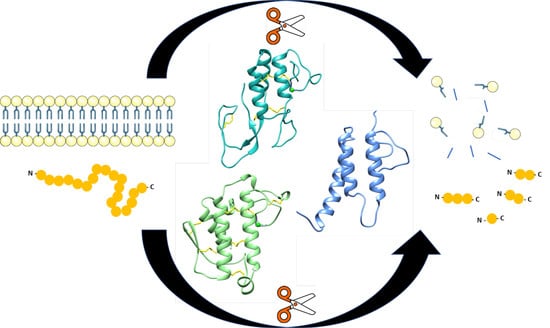
Adeno-Associated Virus VP1u Exhibits Protease Activity
Abstract
:1. Introduction
2. Materials and Methods
2.1. AAV2 Plasmid Mutagenesis
2.2. Production and Purification of rAAV Capsids
2.3. Production and Purification of AAV Virus-Like Particles (VLPs)
2.4. Negative-Stain Transmission Electron Microscopy
2.5. Substrate Preparation and Protease Assays
2.6. PLA2 Assays
2.7. Cellular Transduction Assays
2.8. Structural Modeling and Sequence Alignments
3. Results and Discussion
3.1. AAV Protease Function Is VP1u Dependent, Active at Physiological pH, Calcium Enhanced and Inhibited by a Protein Protease Inhibitor
3.2. The Targets for AAV2 Protease Activity Are Disordered or Unfolded Proteins
3.3. Protease Function May Be a General Activity for PLA2 Enzymes
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gao, G.; Vandenberghe, L.H.; Alvira, M.R.; Lu, Y.; Calcedo, R.; Zhou, X.; Wilson, J.M. Clades of Adeno-Associated Viruses Are Widely Disseminated in Human Tissues. J. Virol. 2004, 78, 6381–6388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Vliet, K.; Mohiuddin, Y.; McClung, S.; Blouin, V.; Rolling, F.; Moullier, P.; Agbandje-McKenna, M.; Snyder, R.O. Adeno-Associated Virus Capsid Serotype Identification: Analytical Methods Development and Application. J. Virol. Methods 2009, 159, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Ylä-Herttuala, S. Endgame: Glybera Finally Recommended for Approval as the First Gene Therapy Drug in the European Union. Mol. Ther. 2012, 20, 1831–1832. [Google Scholar] [CrossRef] [PubMed]
- Smalley, E. First AAV Gene Therapy Poised for Landmark Approval. Nat. Biotechnol. 2017, 35, 998–999. [Google Scholar] [CrossRef]
- Snijder, J.; van de Waterbeemd, M.; Damoc, E.; Denisov, E.; Grinfeld, D.; Bennett, A.; Agbandje-McKenna, M.; Makarov, A.; Heck, A.J.R. Defining the Stoichiometry and Cargo Load of Viral and Bacterial Nanoparticles by Orbitrap Mass Spectrometry. J. Am. Chem. Soc. 2014, 136, 7295–7299. [Google Scholar] [CrossRef] [PubMed]
- Dennis, E.A.; Cao, J.; Hsu, Y.H.; Magrioti, V.; Kokotos, G. Phospholipase A2 Enzymes: Physical Structure, Biological Function, Disease Implication, Chemical Inhibition and Therapeutic Intervention. Chem. Rev. 2011, 111, 6130–6185. [Google Scholar] [CrossRef] [PubMed]
- Zádori, Z.; Szelei, J.; Lacoste, M.-C.; Li, Y.; Gariépy, S.; Raymond, P.; Allaire, M.; Nabi, I.R.; Tijssen, P. A Viral Phospholipase A2 Is Required for Parvovirus Infectivity. Dev. Cell 2001, 1, 291–302. [Google Scholar] [CrossRef] [Green Version]
- Girod, A.; Wobus, C.E.; Zádori, Z.; Ried, M.; Leike, K.; Tijssen, P.; Kleinschmidt, J.A.; Hallek, M. The VP1 Capsid Protein of Adeno-Associated Virus Type 2 Is Carrying a Phospholipase A2 Domain Required for Virus Infectivity. J. Gen. Virol. 2002, 83, 973–978. [Google Scholar] [CrossRef] [PubMed]
- Salganik, M.; Venkatakrishnan, B.; Bennett, A.; Lins, B.; Yarbrough, J.; Muzyczka, N.; Agbandje-McKenna, M.; McKenna, R. Evidence for pH-Dependent Protease Activity in the Adeno-Associated Virus Capsid. J. Virol. 2012, 86, 11877–11885. [Google Scholar] [CrossRef] [Green Version]
- Tong, L. Viral Proteases. Chem. Rev. 2002, 102, 4609–4626. [Google Scholar] [CrossRef]
- Choi, H.-K.; Tong, L.; Minor, W.; Dumas, P.; Boege, U.; Rossmann, M.G.; Wengler, G. Structure of Sindbis Virus Core Protein Reveals a Chymotrypsin-like Serine Proteinase and the Organization of the Virion. Nature 1991, 354, 37–43. [Google Scholar] [CrossRef]
- Rabinowitz, J.E.; Rolling, F.; Li, C.; Conrath, H.; Xiao, W.; Xiao, X.; Samulski, R.J. Cross-Packaging of a Single Adeno-Associated Virus (AAV) Type 2 Vector Genome into Multiple AAV Serotypes Enables Transduction with Broad Specificity. J. Virol. 2002, 76, 791–801. [Google Scholar] [CrossRef] [Green Version]
- Bennett, A.; Patel, S.; Mietzsch, M.; Jose, A.; Lins-Austin, B.; Yu, J.C.; Bothner, B.; McKenna, R.; Agbandje-McKenna, M. Thermal Stability as a Determinant of AAV Serotype Identity. Mol. Ther. Methods Clin. Dev. 2017, 6, 171–182. [Google Scholar] [CrossRef]
- DiMattia, M.; Govindasamy, L.; Levy, H.C.; Gurda-Whitaker, B.; Kalina, A.; Kohlbrenner, E.; Chiorini, J.A.; McKenna, R.; Muzyczka, N.; Zolotukhin, S.; et al. Production, Purification, Crystallization and Preliminary X-Ray Structural Studies of Adeno-Associated Virus Serotype 5. Acta Crystallogr. Sect. F. Struct. Biol. Cryst. Commun. 2005, 61, 917–921. [Google Scholar] [CrossRef]
- Xiao, X.; Li, J.; Samulski, R.J. Production of High-Titer Recombinant Adeno-Associated Virus Vectors in the Absence of Helper Adenovirus. J. Virol. 1998, 72, 2224–2232. [Google Scholar]
- Zolotukhin, S.; Potter, M.; Hauswirth, W.W.; Guy, J.; Muzyczka, N. A “Humanized” Green Fluorescent Protein CDNA Adapted for High-Level Expression in Mammalian Cells. J. Virol. 1996, 70, 4646–4654. [Google Scholar]
- Matsushita, T.; Elliger, S.; Elliger, C.; Podsakoff, G.; Villarreal, L.; Kurtzman, G.; Iwaki, Y.; Colosi, P. Adeno-Associated Virus Vectors Can Be Efficiently Produced without Helper Virus. Gene Ther. 1998, 5, 938–945. [Google Scholar] [CrossRef]
- Zolotukhin, S.; Byrne, B.J.; Mason, E.; Zolotukhin, I.; Potter, M.; Chesnut, K.; Summerford, C.; Samulski, R.J.; Muzyczka, N. Recombinant Adeno-Associated Virus Purification Using Novel Methods Improves Infectious Titer and Yield. Gene Ther. 1999, 6, 973–985. [Google Scholar] [CrossRef]
- Brooke, D.; Movahed, N.; Bothner, B. Universal Buffers for Use in Biochemistry and Biophysical Experiments. AIMS Biophys. 2015, 2, 336–342. [Google Scholar] [CrossRef]
- Mietzsch, M.; Grasse, S.; Zurawski, C.; Weger, S.; Bennett, A.; Agbandje-McKenna, M.; Muzyczka, N.; Zolotukhin, S.; Heilbronn, R. OneBac: Platform for Scalable and High-Titer Production of Adeno-Associated Virus Serotype 1–12 Vectors for Gene Therapy. Hum. Gene Ther. 2014, 25, 212–222. [Google Scholar] [CrossRef]
- Källberg, M.; Wang, H.; Wang, S.; Peng, J.; Wang, Z.; Lu, H.; Xu, J. Template-Based Protein Structure Modeling Using the RaptorX Web Server. Nat. Protoc. 2012, 7, 1511–1522. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a Visualization System for Exploratory Research and Analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- DeLano, W.L. Pymol: An open-source molecular graphics tool. CCP4 Newsl. Protein Crystallogr. 2002, 40, 82–92. [Google Scholar]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Soding, J.; et al. Fast, Scalable Generation of High-Quality Protein Multiple Sequence Alignments Using Clustal Omega. Mol. Syst. Biol. 2014, 7, 539. [Google Scholar] [CrossRef]
- Suikkanen, S.; Antila, M.; Jaatinen, A.; Vihinen-Ranta, M.; Vuento, M. Release of Canine Parvovirus from Endocytic Vesicles. Virology 2003, 316, 267–280. [Google Scholar] [CrossRef]
- Vestling, M.M.; Murphy, C.M.; Fenselau, C. Recognition of Trypsin Autolysis Products by High-Performance Liquid Chromatography and Mass Spectrometry. Anal. Chem. 1990, 62, 2391–2394. [Google Scholar] [CrossRef]
- Vajda, T.; Garai, A. Comparison of the Effect of Calcium(II) and Manganese(II) Ions on Trypsin Autolysis. J. Inorg. Biochem. 1981, 15, 307–315. [Google Scholar] [CrossRef]
- Grieger, J.C.; Johnson, J.S.; Gurda-Whitaker, B.; Agbandje-McKenna, M.; Samulski, R.J. Surface-Exposed Adeno-Associated Virus Vp1-NLS Capsid Fusion Protein Rescues Infectivity of Noninfectious Wild-Type Vp2/Vp3 and Vp3-Only Capsids but Not That of Fivefold Pore Mutant Virions. J. Virol. 2007, 81, 7833–7843. [Google Scholar] [CrossRef]
- Kronenberg, S.; Böttcher, B.; von der Lieth, C.W.; Bleker, S.; Kleinschmidt, J.A. A Conformational Change in the Adeno-Associated Virus Type 2 Capsid Leads to the Exposure of Hidden VP1 N Termini. J. Virol. 2005, 79, 5296–5303. [Google Scholar] [CrossRef] [Green Version]
- Venkatakrishnan, B.; Yarbrough, J.; Domsic, J.; Bennett, A.; Bothner, B.; Kozyreva, O.G.; Samulski, R.J.; Muzyczka, N.; McKenna, R.; Agbandje-McKenna, M. Structure and Dynamics of Adeno-Associated Virus Serotype 1 VP1-Unique N-Terminal Domain and Its Role in Capsid Trafficking. J. Virol. 2013, 87, 4974–4984. [Google Scholar] [CrossRef] [Green Version]
- Nam, H.-J.; Gurda, B.L.; McKenna, R.; Potter, M.; Byrne, B.; Salganik, M.; Muzyczka, N.; Agbandje-McKenna, M. Structural Studies of Adeno-Associated Virus Serotype 8 Capsid Transitions Associated with Endosomal Trafficking. J. Virol. 2011, 85, 11791–11799. [Google Scholar] [CrossRef] [Green Version]
- Canaan, S.; Zádori, Z.; Ghomashchi, F.; Bollinger, J.; Sadilek, M.; Moreau, M.E.; Tijssen, P.; Gelb, M.H. Interfacial Enzymology of Parvovirus Phospholipases A2. J. Biol. Chem. 2004, 279, 14502–14508. [Google Scholar] [CrossRef] [Green Version]
- Gettins, P.G.; Hahn, K.H.; Crews, B.C. Alpha 2-Macroglobulin Bait Region Variants. A Role for the Bait Region in Tetramer Formation. J. Biol. Chem. 1995, 270, 14160–14167. [Google Scholar] [CrossRef]
- Dickson, R.B.; Willingham, M.C.; Pastan, I. Binding and Internalization of 125I-Alpha 2-Macroglobulin by Cultured Fibroblasts. J. Biol. Chem. 1981, 256, 3454–3459. [Google Scholar]
- Ashcom, J.D.; Tiller, S.E.; Dickerson, K.; Cravens, J.L.; Argraves, W.S.; Strickland, D.K. The Human Alpha 2-Macroglobulin Receptor: Identification of a 420-KD Cell Surface Glycoprotein Specific for the Activated Conformation of Alpha 2-Macroglobulin. J. Cell Biol. 1990, 110, 1041–1048. [Google Scholar] [CrossRef]
- van der Valk, J.; Brunner, D.; De Smet, K.; Fex Svenningsen, Å.; Honegger, P.; Knudsen, L.E.; Lindl, T.; Noraberg, J.; Price, A.; Scarino, M.L.; et al. Optimization of Chemically Defined Cell Culture Media – Replacing Fetal Bovine Serum in Mammalian in Vitro Methods. Toxicol. In Vitro 2010, 24, 1053–1063. [Google Scholar] [CrossRef]
- Wang, M.; Sun, J.; Crosby, A.; Woodard, K.; Hirsch, M.L.; Samulski, R.J.; Li, C. Direct Interaction of Human Serum Proteins with AAV Virions to Enhance AAV Transduction: Immediate Impact on Clinical Applications. Gene Ther. 2017, 24, 49–59. [Google Scholar] [CrossRef]
- Horne, D.S. Casein Structure, Self-Assembly and Gelation. Curr. Opin. Colloid Interface Sci. 2002, 7, 456–461. [Google Scholar] [CrossRef]
- Badii, F.; Howell, N.K. Fish Gelatin: Structure, Gelling Properties and Interaction with Egg Albumen Proteins. Food Hydrocoll. 2006, 20, 630–640. [Google Scholar] [CrossRef]
- Takeda, K.; Wada, A.; Yamamoto, K.; Moriyama, Y.; Aoki, K. Conformational Change of Bovine Serum Albumin by Heat Treatment. J. Protein Chem. 1989, 8, 653–659. [Google Scholar] [CrossRef]
- Van der Lee, R.; Buljan, M.; Lang, B.; Weatheritt, R.J.; Daughdrill, G.W.; Dunker, A.K.; Fuxreiter, M.; Gough, J.; Gsponer, J.; Jones, D.T.; et al. Classification of Intrinsically Disordered Regions and Proteins. Chem. Rev. 2014, 114, 6589–6631. [Google Scholar] [CrossRef] [Green Version]
- Denning, D.P.; Patel, S.S.; Uversky, V.; Fink, A.L.; Rexach, M. Disorder in the Nuclear Pore Complex: The FG Repeat Regions of Nucleoporins Are Natively Unfolded. Proc. Natl. Acad. Sci. USA 2003, 100, 2450–2455. [Google Scholar] [CrossRef]
- Porwal, M.; Cohen, S.; Snoussi, K.; Popa-Wagner, R.; Anderson, F.; Dugot-Senant, N.; Wodrich, H.; Dinsart, C.; Kleinschmidt, J.A.; Panté, N.; et al. Parvoviruses Cause Nuclear Envelope Breakdown by Activating Key Enzymes of Mitosis. PLoS Pathog. 2013, 9, e1003671. [Google Scholar] [CrossRef]
- Cavigiolio, G.; Jayaraman, S. Proteolysis of Apolipoprotein A-I by Secretory Phospholipase A2: A New Link between Inflammation and Atherosclerosis. J. Biol. Chem. 2014, 289, 10011–10023. [Google Scholar] [CrossRef]
- Wu, P.; Xiao, W.; Conlon, T.; Hughes, J.; Agbandje-McKenna, M.; Ferkol, T.; Flotte, T.; Muzyczka, N. Mutational Analysis of the Adeno-Associated Virus Type 2 (AAV2) Capsid Gene and Construction of AAV2 Vectors with Altered Tropism. J. Virol. 2000, 74, 8635–8647. [Google Scholar] [CrossRef] [Green Version]
- Rebek, J. On the Structure of Histidine and Its Role in Enzyme Active Sites. Struct. Chem. 1990, 1, 129–131. [Google Scholar] [CrossRef]
- Ekici, O.D.; Paetzel, M.; Dalbey, R.E. Unconventional Serine Proteases: Variations on the Catalytic Ser/His/Asp Triad Configuration. Protein Sci. 2008, 17, 2023–2037. [Google Scholar] [CrossRef]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kurian, J.J.; Lakshmanan, R.; Chmely, W.M.; Hull, J.A.; Yu, J.C.; Bennett, A.; McKenna, R.; Agbandje-McKenna, M. Adeno-Associated Virus VP1u Exhibits Protease Activity. Viruses 2019, 11, 399. https://doi.org/10.3390/v11050399
Kurian JJ, Lakshmanan R, Chmely WM, Hull JA, Yu JC, Bennett A, McKenna R, Agbandje-McKenna M. Adeno-Associated Virus VP1u Exhibits Protease Activity. Viruses. 2019; 11(5):399. https://doi.org/10.3390/v11050399
Chicago/Turabian StyleKurian, Justin J., Renuk Lakshmanan, William M. Chmely, Joshua A. Hull, Jennifer C. Yu, Antonette Bennett, Robert McKenna, and Mavis Agbandje-McKenna. 2019. "Adeno-Associated Virus VP1u Exhibits Protease Activity" Viruses 11, no. 5: 399. https://doi.org/10.3390/v11050399