Characterization of Neutralizing Monoclonal Antibodies and Identification of a Novel Conserved C-Terminal Linear Epitope on the Hemagglutinin Protein of the H9N2 Avian Influenza Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Proteins, and Reagents
2.2. Mouse Immunization and Generation of Monoclonal Antibodies
2.3. IPMA Test
2.4. Indirect ELISA
2.5. Microneutralization Assay
2.6. Hemagglutination Inhibition Assay
2.7. Western Blotting
2.8. Dot Blotting
2.9. Mapping the Linear B Cell Epitope of the Recombinant HA Protein
2.10. Structural Locations of Positive Peptides
2.11. Conservation Analysis
3. Results
3.1. H9N2 HA Immunization and Generation of mAbs
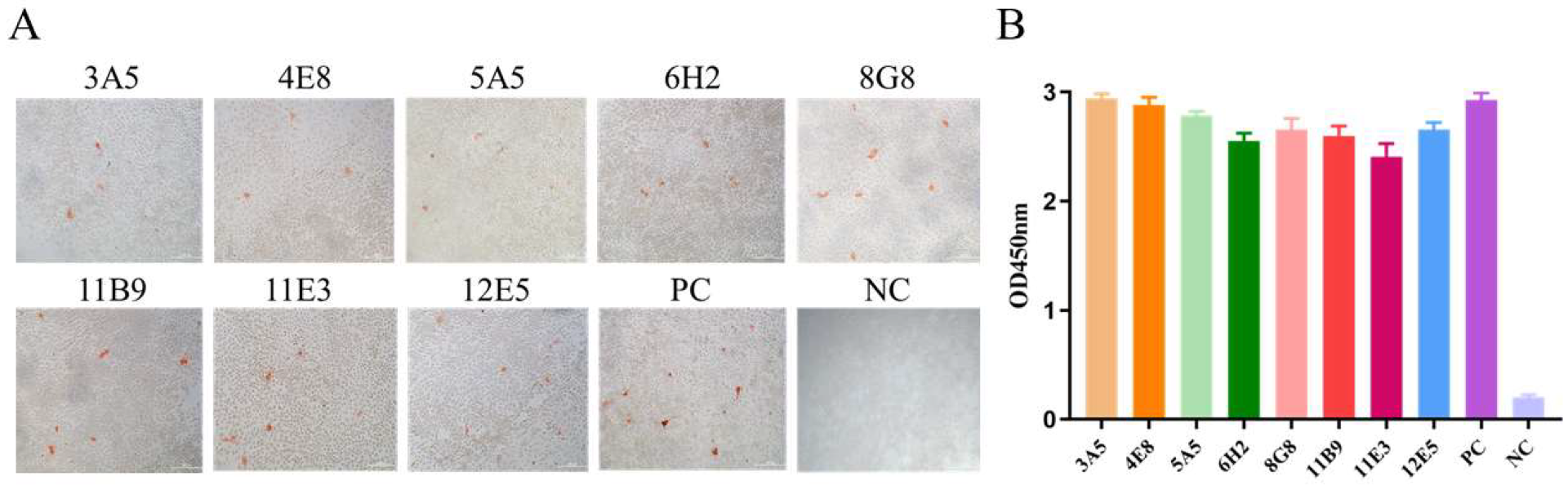
3.2. mAbs Screening
3.3. Characterization of the Generated mAbs
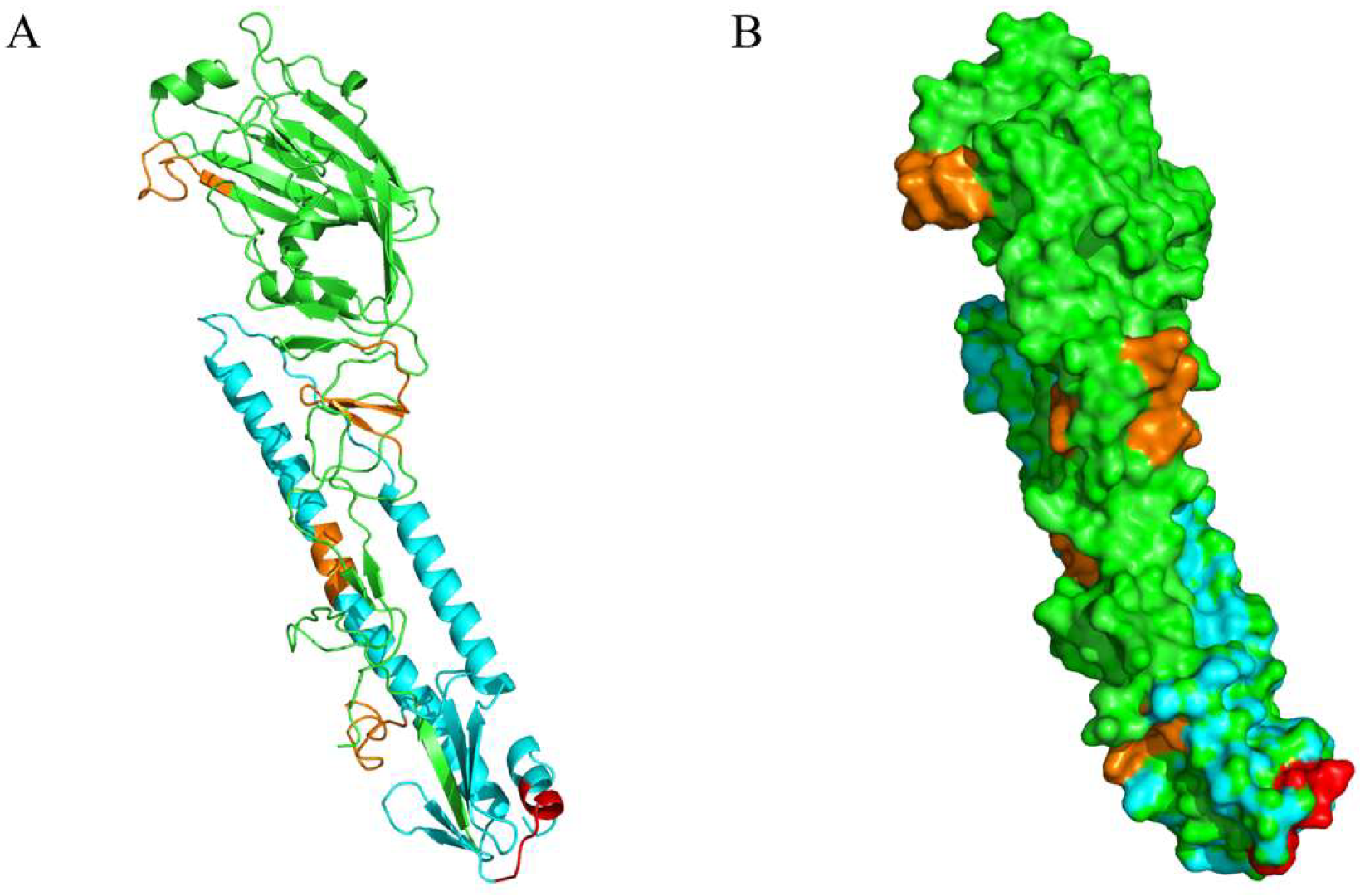
3.4. Mapping of B Cell Epitope on the HA Protein
3.5. Biological Data Analysis
3.6. Conservation Analysis of the Novel Epitope
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Eladl, A.H.; Mosad, S.M.; El-Shafei, R.A.; Saleh, R.M.; Ali, H.S.; Badawy, B.M.; Elshal, M.F. Immunostimulant effect of a mixed herbal extract on infectious bursal disease virus (IBDV) vaccinated chickens in the context of a co-infection model of avian influenza virus H9N2 and IBDV. Comp. Immunol. Microbiol. Infect. Dis. 2020, 72, 101505. [Google Scholar] [CrossRef] [PubMed]
- Arafat, N.; El Rahman, S.A.; Naguib, D.; El-Shafei, R.A.; Abdo, W.; Eladl, A.H. Co-infection of Salmonella enteritidis with H9N2 avian influenza virus in chickens. Avian Pathol. 2020, 49, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Arafat, N.; Eladl, A.H.; Marghani, B.H.; Saif, M.A.; El-Shafei, R. Enhanced infection of avian influenza virus H9N2 with infectious laryngeotracheitis vaccination in chickens. Veter-Microbiol. 2018, 219, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; James, J.; Sealy, J.E.; Iqbal, M. A Global Perspective on H9N2 Avian Influenza Virus. Viruses 2019, 11, 620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.-T.; Linster, M.; Mendenhall, I.H.; Su, Y.C.F.; Smith, G.J.D. Avian influenza viruses in humans: Lessons from past outbreaks. Br. Med. Bull. 2019, 132, 81–95. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, X.; Zhou, J.; Shi, S.; Shen, T.; Chen, L.; Zhang, M.; Liao, C.; Wang, C. Development of PDA Nanoparticles for H9N2 Avian Influenza BPP-V/BP-IV Epitope Peptide Vaccines: Immunogenicity and Delivery Efficiency Improvement. Front. Immunol. 2021, 12, 693972. [Google Scholar] [CrossRef]
- Awadin, W.F.; Eladl, A.H.; El-Shafei, R.A.; El-Adl, M.A.; Aziza, A.E.; Ali, H.S.; Saif, M.A. Effect of omega-3 rich diet on the response of Japanese quails (Coturnix coturnix japonica) infected with Newcastle disease virus or avian influenza virus H9N2. Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 2019, 228, 108668. [Google Scholar] [CrossRef]
- Bi, Y.; Li, J.; Li, S.; Fu, G.; Jin, T.; Zhang, C.; Yang, Y.; Ma, Z.; Tian, W.; Li, J.; et al. Dominant subtype switch in avian influenza viruses during 2016–2019 in China. Nat. Commun. 2020, 11, 5909. [Google Scholar] [CrossRef]
- Chen, H.; Yuan, H.; Gao, R.; Zhang, J.; Wang, D.; Xiong, Y.; Fan, G.; Yang, F.; Li, X.; Zhou, J.; et al. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: A descriptive study. Lancet 2014, 383, 714–721. [Google Scholar] [CrossRef]
- Pu, J.; Wang, S.; Yin, Y.; Zhang, G.; Carter, R.A.; Wang, J.; Xu, G.; Sun, H.; Wang, M.; Wen, C.; et al. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proc. Natl. Acad. Sci. USA 2015, 112, 548–553. [Google Scholar] [CrossRef]
- Liu, D.; Shi, W.; Gao, G.F. Poultry carrying H9N2 act as incubators for novel human avian influenza viruses. Lancet 2014, 383, 869. [Google Scholar] [CrossRef]
- Luo, M. Viral Molecular Machines; Springer US: Boston, MA, USA, 2012; pp. 201–221. [Google Scholar] [CrossRef]
- Boonsathorn, N.; Panthong, S.; Koksunan, S.; Chittaganpitch, M.; Phuygun, S.; Waicharoen, S.; Prachasupap, A.; Sasaki, T.; Kubota-Koketsu, R.; Yasugi, M.; et al. A human monoclonal antibody derived from a vaccinated volunteer recognizes heterosubtypically a novel epitope on the hemagglutinin globular head of H1 and H9 influenza A viruses. Biochem. Biophys. Res. Commun. 2014, 452, 865–870. [Google Scholar] [CrossRef] [Green Version]
- Krause, J.C.; Tsibane, T.; Tumpey, T.M.; Huffman, C.J.; Basler, C.F.; Crowe, J.E. A Broadly Neutralizing Human Monoclonal Antibody That Recognizes a Conserved, Novel Epitope on the Globular Head of the Influenza H1N1 Virus Hemagglutinin. J. Virol. 2011, 85, 10905–10908. [Google Scholar] [CrossRef] [Green Version]
- Ekiert, D.C.; Kashyap, A.K.; Steel, J.; Rubrum, A.; Bhabha, G.; Khayat, R.; Lee, J.H.; Dillon, M.A.; O’Neil, R.E.; Faynboym, A.M.; et al. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 2012, 489, 526–532. [Google Scholar] [CrossRef] [Green Version]
- Tan, G.S.; Leon, P.E.; Albrecht, R.A.; Margine, I.; Hirsh, A.; Bahl, J.; Krammer, F. Broadly-Reactive Neutralizing and Non-neutralizing Antibodies Directed against the H7 Influenza Virus Hemagglutinin Reveal Divergent Mechanisms of Protection. PLOS Pathog. 2016, 12, e1005578. [Google Scholar] [CrossRef]
- Iba, Y.; Fujii, Y.; Ohshima, N.; Sumida, T.; Kubota-Koketsu, R.; Ikeda, M.; Wakiyama, M.; Shirouzu, M.; Okada, J.; Okuno, Y.; et al. Conserved Neutralizing Epitope at Globular Head of Hemagglutinin in H3N2 Influenza Viruses. J. Virol. 2014, 88, 7130–7144. [Google Scholar] [CrossRef] [Green Version]
- Yoshida, R.; Igarashi, M.; Ozaki, H.; Kishida, N.; Tomabechi, D.; Kida, H.; Ito, K.; Takada, A. Cross-Protective Potential of a Novel Monoclonal Antibody Directed against Antigenic Site B of the Hemagglutinin of Influenza A Viruses. PLoS Pathog. 2009, 5, e1000350. [Google Scholar] [CrossRef] [Green Version]
- Ekiert, D.C.; Bhabha, G.; Elsliger, M.-A.; Friesen, R.H.E.; Jongeneelen, M.; Throsby, M.; Goudsmit, J.; Wilson, I.A. Antibody Recognition of a Highly Conserved Influenza Virus Epitope. Science 2009, 324, 246–251. [Google Scholar] [CrossRef] [Green Version]
- Sui, J.; Hwang, W.C.; Perez, S.; Wei, G.; Aird, D.; Chen, L.-M.; Santelli, E.; Stec, B.; Cadwell, G.; Ali, M.; et al. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat. Struct. Mol. Biol. 2009, 16, 265–273. [Google Scholar] [CrossRef]
- Corti, D.; Voss, J.; Gamblin, S.J.; Codoni, G.; Macagno, A.; Jarrossay, D.; Vachieri, S.G.; Pinna, D.; Minola, A.; Vanzetta, F.; et al. A Neutralizing Antibody Selected from Plasma Cells That Binds to Group 1 and Group 2 Influenza A Hemagglutinins. Science 2011, 333, 850–856. [Google Scholar] [CrossRef]
- Dreyfus, C.; Laursen, N.S.; Kwaks, T.; Zuijdgeest, D.; Khayat, R.; Ekiert, D.C.; Lee, J.H.; Metlagel, Z.; Bujny, M.V.; Jongeneelen, M.; et al. Highly conserved protective epitopes on influenza B viruses. Science 2012, 337, 1343–1348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Ma, M.L.; Lei, Q.; Wang, F.; Hong, W.; Lai, D.Y.; Hou, H.; Xu, Z.W.; Zhang, B.; Chen, H.; et al. Linear epitope landscape of the SARS-CoV-2 Spike protein constructed from 1051 COVID-19 patients. Cell Rep. 2021, 34, 108915. [Google Scholar] [CrossRef] [PubMed]
- Takemasa, E.; Liu, S.; Hasegawa, H. Production of Neutralizing Antibody. Methods Mol Biol. 2018, 1868, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Tijssen, P. Practice and Theory of Enzyme Immunoassays; Elsevier Science Ltd.: Amsterdam, The Netherlands, 1985; pp. 173–210. [Google Scholar]
- Beatty, J.; Beatty, B.G.; Vlahos, W.G. Measurement of monoclonal antibody affinity by non-competitive enzyme immunoassay. J. Immunol. Methods 1987, 100, 173–179. [Google Scholar] [CrossRef]
- Standard Operating Procedures for the National Influenza Center. Available online: https://ivdc.chinacdc.cn/cnic/en/Surveillance/sgs/201606/t20160602_130534.htm (accessed on 30 March 2007).
- Liu, Y.; Wang, J.; Chen, Y.; Wang, A.; Wei, Q.; Yang, S.; Feng, H.; Chai, S.; Liu, D.; Zhang, G. Identification of a dominant linear epitope on the VP2 capsid protein of porcine parvovirus and characterization of two monoclonal antibodies with neutralizing abilities. Int. J. Biol. Macromol. 2020, 163, 2013–2022. [Google Scholar] [CrossRef]
- Xu, Q.; Guo, J.; Ma, F.; Liu, L.; Wang, Y.; Zhang, S.; Niu, X.; Li, X.; Jiang, M.; Wang, Y.; et al. A novel linear epitope at the C-terminal region of the classical swine fever virus E2 protein elicits neutralizing activity. Int. J. Biol. Macromol. 2021, 189, 837–846. [Google Scholar] [CrossRef]
- Chen, H.; Subbarao, K.; Swayne, D.; Chen, Q.; Lu, X.; Katz, J.; Cox, N.; Matsuoka, Y. Generation and evaluation of a high-growth reassortant H9N2 influenza A virus as a pandemic vaccine candidate. Vaccine 2003, 21, 1974–1979. [Google Scholar] [CrossRef]
- Parvin, R.; Heenemann, K.; Halami, M.Y.; Chowdhury, E.H.; Islam, M.R.; Vahlenkamp, T.W. Full-genome analysis of avian influenza virus H9N2 from Bangladesh reveals internal gene reassortments with two distinct highly pathogenic avian influenza viruses. Arch. Virol. 2014, 159, 1651–1661. [Google Scholar] [CrossRef]
- Guan, Y.; Shortridge, K.F.; Krauss, S.; Webster, R.G. Molecular characterization of H9N2 influenza viruses: Were they the donors of the “internal” genes of H5N1 viruses in Hong Kong? Proc. Natl. Acad. Sci. USA 1999, 96, 9363–9367. [Google Scholar] [CrossRef]
- Awadin, W.F.; Eladl, A.H.; El-Shafei, R.; El-Adl, M.; Ali, H.S. Immunological and pathological effects of vitamin E with Fetomune Plus® on chickens experimentally infected with avian influenza virus H9N2. Veter. Microbiol. 2019, 231, 24–32. [Google Scholar] [CrossRef]
- Shi, S.H.; Yang, W.T.; Yang, G.L.; Cong, Y.L.; Huang, H.B.; Wang, Q.; Cai, R.P.; Ye, L.P.; Hu, J.T.; Zhou, J.Y.; et al. Immunoprotection against influenza virus H9N2 by the oral administration of recombinant Lactobacillus plantarumNC8 expressing hemagglutinin in BALB/c mice. Virology 2014, 464–465, 166–176. [Google Scholar] [CrossRef] [Green Version]
- Eladl, A.H.; Arafat, N.; El-Shafei, R.A.; Farag, V.M.; Saleh, R.M.; Awadin, W.F. Comparative immune response and pathogenicity of the H9N2 avian influenza virus after administration of Immulant®, based on Echinacea and Nigella sativa, in stressed chickens. Comp. Immunol. Microbiol. Infect. Dis. 2019, 65, 165–175. [Google Scholar] [CrossRef]
- Eladl, A.H.; Alzayat, A.A.; Ali, H.S.; Fahmy, H.A.; Ellakany, H.F. Comparative molecular characterization, pathogenicity and seroprevalence of avian influenza virus H9N2 in commercial and backyard poultry flocks. Comp. Immunol. Microbiol. Infect. Dis. 2019, 64, 81–89. [Google Scholar] [CrossRef]
- Lukosaityte, D.; Sadeyen, J.-R.; Shrestha, A.; Sealy, J.E.; Bhat, S.; Chang, P.; Digard, P.; Iqbal, M. Engineered Recombinant Single Chain Variable Fragment of Monoclonal Antibody Provides Protection to Chickens Infected with H9N2 Avian Influenza. Vaccines 2020, 8, 118. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Y.; Yang, F.; Liu, F.; Yao, H.; Wu, N.; Wu, H. Antigen-capture ELISA and immunochromatographic test strip to detect the H9N2 subtype avian influenza virus rapidly based on monoclonal antibodies. Virol. J. 2021, 18, 1–10. [Google Scholar] [CrossRef]
- Wang, F.; Wu, J.; Wang, Y.; Wan, Z.; Shao, H.; Qian, K.; Ye, J.; Qin, A. Identification of key residues involved in the neuraminidase antigenic variation of H9N2 influenza virus. Emerg. Microbes Infect. 2021, 10, 210–219. [Google Scholar] [CrossRef]
- Russell, C.A.; Jones, T.C.; Barr, I.G.; Cox, N.J.; Garten, R.J.; Gregory, V.; Gust, I.D.; Hampson, A.W.; Hay, A.J.; Hurt, A.C.; et al. The Global Circulation of Seasonal Influenza A (H3N2) Viruses. Science 2008, 320, 340–346. [Google Scholar] [CrossRef]
- Margolin, E.; Chapman, R.; Williamson, A.-L.; Rybicki, E.P.; Meyers, A.E. Production of complex viral glycoproteins in plants as vaccine immunogens. Plant Biotechnol. J. 2018, 16, 1531–1545. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Shen, T.; Zhou, J.; Chen, L.; Shi, S.; Wang, X.; Zhang, M.; Wang, C.; Liao, C. Bursal peptide BP-IV as a novel immunoadjuvant enhances the protective efficacy of an epitope peptide vaccine containing T and B cell epitopes of the H9N2 avian influenza virus. Microb. Pathog. 2021, 158, 105095. [Google Scholar] [CrossRef]
- Rhee, J.W.; Kim, D.; Park, B.K.; Kwon, S.; Cho, S.; Lee, I.; Park, M.S.; Seo, J.N.; Kim, Y.S.; Choi, H.S.; et al. Immunization with a hemagglutinin-derived synthetic peptide formulated with a CpG-DNA-liposome complex induced protection against lethal influenza virus infection in mice. PLoS ONE 2012, 7, e48750. [Google Scholar] [CrossRef] [Green Version]
- Chun, S.; Li, C.; Van Domselaar, G.; Wang, J.; Farnsworth, A.; Cui, X.; Rode, H.; Cyr, T.D.; He, R.; Li, X. Universal antibodies and their applications to the quantitative determination of virtually all subtypes of the influenza A viral hemagglutinins. Vaccine 2008, 26, 6068–6076. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Huang, N.; Fan, M.; Zhao, L.; Luo, Y.; Ding, P.; Tian, M.; Liu, Q.; Guo, Y.; Zhao, J.; et al. Hemagglutinin stalk-based monoclonal antibody elicits broadly reactivity against group 1 influenza A virus. Virol. J. 2020, 17, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Yang, D.; Ren, Q.; Yang, Y.; Liu, X.; Xu, X.; Liu, W.; Chen, S.; Peng, D.; Liu, X. Identification and characterization of a novel antigenic epitope in the hemagglutinin of the escape mutants of H9N2 avian influenza viruses. Veter. Microbiol. 2015, 178, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Gui, M.; Xiang, Y. Structural intermediates in the low pH-induced transition of influenza hemagglutinin. PLoS Pathog. 2020, 16, e1009062. [Google Scholar] [CrossRef]
- Ren, H.; Zhou, P. Epitope-focused vaccine design against influenza A and B viruses. Curr. Opin. Immunol. 2016, 42, 83–90. [Google Scholar] [CrossRef] [Green Version]
- Blijleven, J.S.; Boonstra, S.; Onck, P.R.; van der Giessen, E.; van Oijen, A.M. Mechanisms of influenza viral membrane fusion. Semin. Cell Dev. Biol. 2016, 60, 78–88. [Google Scholar] [CrossRef] [Green Version]
- Boonstra, S.; Blijleven, J.S.; Roos, W.H.; Onck, P.R.; van der Giessen, E.; van Oijen, A.M. Hemagglutinin-Mediated Membrane Fusion: A Biophysical Perspective. Annu. Rev. Biophys. 2018, 47, 153–173. [Google Scholar] [CrossRef]
- Wild, C.T.; Shugars, D.C.; Greenwell, T.K.; McDanal, C.B.; Matthews, T.J. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 1994, 91, 9770–9774. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.K.; Pessi, A.; Gui, L.; Santoprete, A.; Talekar, A.; Moscona, A.; Porotto, M. Capturing a Fusion Intermediate of Influenza Hemagglutinin with a Cholesterol-conjugated Peptide, a New Antiviral Strategy for Influenza Virus. J. Biol. Chem. 2011, 286, 42141–42149. [Google Scholar] [CrossRef]




| Name | Peptide | Length (aa) |
|---|---|---|
| P1 | CDKICIGYQSTNSTETVDTL | 20 |
| P2 | CTETVDTLTENNVPVTHAKE | 20 |
| P3 | CPVTHAKELLHTEHNGMLCA | 20 |
| P4 | CHNGMLCATNLGHPLILDTC | 20 |
| P5 | CPLILDTCTIEGLIYGNPSC | 20 |
| P6 | CIYGNPSCDLLLGGREWSYI | 20 |
| P7 | CGREWSYIVERPSAVNGLCY | 20 |
| P8 | CAVNGLCYPGNVENLEELRS | 20 |
| P9 | CNLEELRSLFSSARSYQRIQ | 20 |
| P10 | CRSYQRIQIFPDTIWNVSYS | 20 |
| P11 | CIWNVSYSGTSRACSDSFYR | 20 |
| P12 | CSDSFYRSMRWLTQKNNAYP | 20 |
| P13 | CQKNNAYPVQDAQYTNNRGK | 20 |
| P14 | CYTNNRGKNILFMWGINHPP | 20 |
| P15 | CWGINHPPTDTAQTNLYTRT | 20 |
| P16 | CTNLYTRTDTTTSVATEDIN | 20 |
| P17 | CVATEDINRTFKPLIGPRPL | 20 |
| P18 | CLIGPRPLVNGLQGRIDYYW | 20 |
| P19 | CGRIDYYWSVLKPGQTLRVK | 20 |
| P20 | CGQTLRVKSNGNLIAPWYGH | 20 |
| P21 | CIAPWYGHILSGESHGRILK | 20 |
| P22 | CSHGRILKTDLNSGNCVVQC | 20 |
| P23 | CGNCVVQCQTERGGLNTTLP | 20 |
| P24 | CGLNTTLPFHNVSKYAFGNC | 20 |
| P25 | CKYAFGNCPKYVGVKSLKLA | 20 |
| P26 | CVKSLKLAVGLRNVPARSSR | 20 |
| P27 | CVPARSSRGLFGAIAGFIEG | 20 |
| P28 | CIAGFIEGGWSGLVAGWYGF | 20 |
| P29 | CVAGWYGFQHSNDQGVGMAA | 20 |
| P30 | CQGVGMAADRDSTQKAIDKI | 20 |
| P31 | CQKAIDKITSKVNNIVDKMN | 20 |
| P32 | CNIVDKMNKQYEIIDHEFSE | 20 |
| P33 | CIDHEFSEVETRLNMINNKI | 20 |
| P34 | CNMINNKIDDQIQDIWAYNA | 20 |
| P35 | CDIWAYNAELLVLLENQKTL | 20 |
| P36 | CLENQKTLDEHDANVNNLYN | 20 |
| P37 | CNVNNLYNKVKRALGSNAVE | 20 |
| P38 | CLGSNAVEDGKGCFELYHKC | 20 |
| P39 | CFELYHKCDDQCMETIRNGT | 20 |
| P40 | CETIRNGTYNRRKYKEESRL | 20 |
| P41 | CYKEESRLERQKIEGVKLESEGT | 23 |
| Name | Peptide | Length (aa) |
|---|---|---|
| P39-1 | LGSNAVEDGKGC | 12 |
| P39-2 | FELYHKC | 7 |
| P39-3 | DDQCM | 5 |
| P39-4 | CETIRNGT | 8 |
| P39-5 | HKCDDQCM | 8 |
| NO. | mAbs | Types | IPMA Titer | ELISA Titer | Ka (mol/L) |
|---|---|---|---|---|---|
| 1 | 3A5 | IgG1, Kappa | 51,200 | 102,400 | 2.49 × 105 |
| 2 | 4E8 | IgG1, Kappa | 12,800 | 51,200 | 4.17 × 104 |
| 3 | 5A5 | IgG2a, Kappa | 6400 | 12,800 | 2.83 × 104 |
| 4 | 6H2 | IgG1, Kappa | 12,800 | 102,400 | 3.49 × 104 |
| 5 | 8G8 | IgG1, Kappa | 102,400 | 204,800 | 1.21 × 104 |
| 6 | 11B9 | IgG2b, Kappa | 25,600 | 51,200 | 6.7 × 103 |
| 7 | 11E3 | IgG1, Kappa | 12,800 | 51,200 | 7.8 × 103 |
| 8 | 12E5 | IgG1, Kappa | 102,400 | 102,400 | 1.1 × 104 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Li, X.; Xu, Q.; Niu, X.; Zhang, S.; Qu, X.; Chu, H.; Chen, J.; Shi, Q.; Zhang, E.; et al. Characterization of Neutralizing Monoclonal Antibodies and Identification of a Novel Conserved C-Terminal Linear Epitope on the Hemagglutinin Protein of the H9N2 Avian Influenza Virus. Viruses 2022, 14, 2530. https://doi.org/10.3390/v14112530
Wang Y, Li X, Xu Q, Niu X, Zhang S, Qu X, Chu H, Chen J, Shi Q, Zhang E, et al. Characterization of Neutralizing Monoclonal Antibodies and Identification of a Novel Conserved C-Terminal Linear Epitope on the Hemagglutinin Protein of the H9N2 Avian Influenza Virus. Viruses. 2022; 14(11):2530. https://doi.org/10.3390/v14112530
Chicago/Turabian StyleWang, Yanan, Xueyang Li, Qianru Xu, Xiangxiang Niu, Shenli Zhang, Xiaotian Qu, Hongyan Chu, Jinxuan Chen, Qianqian Shi, Erqin Zhang, and et al. 2022. "Characterization of Neutralizing Monoclonal Antibodies and Identification of a Novel Conserved C-Terminal Linear Epitope on the Hemagglutinin Protein of the H9N2 Avian Influenza Virus" Viruses 14, no. 11: 2530. https://doi.org/10.3390/v14112530




