Fifty Shades of Erns: Innate Immune Evasion by the Viral Endonucleases of All Pestivirus Species
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Reagents
2.2. Expression and Purification of Strep-Tagged Erns
2.3. RNase Activity Assay
2.4. Mx Assay
2.5. Western Blot
2.6. Coomassie Staining
2.7. Immunofluorescence Microscopy
3. Results
3.1. Protein Expression and Molecular Weight Determination
3.2. RNase Activity
3.3. Intracellular Localization
3.4. Inhibition of Interferon Expression
4. Discussion
4.1. Protein Purification
4.2. RNase Activity
4.3. Intracellular Localization and IFN Antagonism
4.4. Erns Dimerization
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Smith, D.B.; Meyers, G.; Bukh, J.; Gould, E.A.; Monath, T.; Scott Muerhoff, A.; Pletnev, A.; Rico-Hesse, R.; Stapleton, J.T.; Simmonds, P.; et al. Proposed revision to the taxonomy of the genus Pestivirus, family Flaviviridae. J. Gen. Virol. 2017, 98, 2106–2112. [Google Scholar] [CrossRef]
- King, A.M.Q.; Lefkowitz, E.J.; Mushegian, A.R.; Adams, M.J.; Dutilh, B.E.; Gorbalenya, A.E.; Harrach, B.; Harrison, R.L.; Junglen, S.; Knowles, N.J.; et al. Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch. Virol. 2018, 163, 2601–2631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyers, G.; Ege, A.; Fetzer, C.; von Freyburg, M.; Elbers, K.; Carr, V.; Prentice, H.; Charleston, B.; Schürmann, E.-M. Bovine viral diarrhea virus: Prevention of persistent fetal infection by a combination of two mutations affecting Erns RNase and Npro protease. J. Virol. 2007, 81, 3327–3338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magouras, I.; Mätzener, P.; Rümenapf, T.; Peterhans, E.; Schweizer, M. RNase-dependent inhibition of extra-, but not intracellular, dsRNA-induced IFN synthesis by Erns of pestiviruses. J. Gen. Virol. 2008, 89, 2501–2506. [Google Scholar] [CrossRef] [PubMed]
- Schweizer, M.; Peterhans, E. Pestiviruses. Annu. Rev. Anim. Biosci. 2014, 2, 141–163. [Google Scholar] [CrossRef]
- Tautz, N.; Tews, B.A.; Meyers, G. The molecular biology of pestiviruses. Adv. Virus Res. 2015, 93, 47–160. [Google Scholar] [CrossRef]
- Lussi, C.; Schweizer, M. What can pestiviral endonucleases teach us about innate immunotolerance? Cytokine Growth Factor Rev. 2016, 29, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Rümenapf, T.; Unger, G.; Strauss, J.H.; Thiel, H.J. Processing of the envelope glycoproteins of pestiviruses. J. Virol. 1993, 67, 3288–3294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zürcher, C.M.; Sauter, K.-S.; Mathys, V.; Wyss, F.; Schweizer, M. Prolonged activity of the pestiviral RNase Erns as an interferon antagonist after uptake by clathrin-mediated endocytosis. J. Virol. 2014, 88, 7235–7243. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, R.; Unger, G.; Stark, R.; Schneider-Scherzer, E.; Thiel, H.-J. Identification of a structural glycoprotein of an RNA virus as a ribonuclease. Science 1993, 261, 1169–1171. [Google Scholar] [CrossRef]
- Windisch, J.M.; Schneider, R.; Stark, R.; Weiland, E.; Meyers, G.; Thiel, H.J. RNase of classical swine fever virus: Biochemical characterization and inhibition by virus-neutralizing monoclonal antibodies. J. Virol. 1996, 70, 352–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iqbal, M.; Poole, E.; Goodbourn, S.; McCauley, J.W. Role for bovine viral diarrhea virus Erns glycoprotein in the control of activation of beta interferon by double-stranded RNA. J. Virol. 2004, 78, 136–145. [Google Scholar] [CrossRef] [Green Version]
- Lussi, C.; Schweizer, M. The pestiviral IFN antagonist Erns cleaves dsRNA as nicking endoribonuclease. Cytokine 2015, 76, 81–82. [Google Scholar] [CrossRef]
- Mätzener, P.; Magouras, I.; Rümenapf, T.; Peterhans, E.; Schweizer, M. The viral RNase Erns prevents IFN type-I triggering by pestiviral single- and double-stranded RNAs. Virus Res. 2009, 140, 15–23. [Google Scholar] [CrossRef]
- Irie, M. Structure-function relationships of acid ribonucleases: Lysosomal, vacuolar, and periplasmic enzymes. Pharmacol. Ther. 1999, 81, 77–89. [Google Scholar] [CrossRef]
- Fetzer, C.; Tews, B.A.; Meyers, G. The carboxy-terminal sequence of the pestivirus glycoprotein Erns represents an unusual type of membrane anchor. J. Virol. 2005, 79, 11901–11913. [Google Scholar] [CrossRef] [Green Version]
- Tews, B.A.; Meyers, G. The pestivirus glycoprotein Erns is anchored in plane in the membrane via an amphipathic helix. J. Biol. Chem. 2007, 282, 32730–32741. [Google Scholar] [CrossRef] [Green Version]
- Aberle, D.; Muhle-Goll, C.; Bürck, J.; Wolf, M.; Reißer, S.; Luy, B.; Wenzel, W.; Ulrich, A.S.; Meyers, G. Structure of the membrane anchor of pestivirus glycoprotein Erns, a long tilted amphipathic helix. PLoS Pathog. 2014, 10, e1003973. [Google Scholar] [CrossRef] [PubMed]
- Aberle, D.; Oetter, K.-M.; Meyers, G. Lipid binding of the amphipathic helix serving as membrane anchor of pestivirus glycoprotein Erns. PLoS ONE 2015, 10, e0135680. [Google Scholar] [CrossRef] [Green Version]
- Langedijk, J.P.M.; van Veelen, P.A.; Schaaper, W.M.M.; de Ru, A.H.; Meloen, R.H.; Hulst, M.M. A structural model of pestivirus Erns based on disulfide bond connectivity and homology modeling reveals an extremely rare vicinal disulfide. J. Virol. 2002, 76, 10383–10392. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, M.; Flick-Smith, H.; McCauley, J.W. Interactions of bovine viral diarrhoea virus glycoprotein Erns with cell surface glycosaminoglycans. J. Gen. Virol. 2000, 81, 451–459. [Google Scholar] [CrossRef]
- Hulst, M.M.; van Gennip, R.; Vlot, A.C.; Schooten, E.; de Smit, A.J.; Moormann, R.J.M. Interaction of classical swine fever virus with membrane-associated heparan sulfate: Role for virus replication in vivo and virulence. J. Virol. 2001, 75, 9585–9595. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, M.; McCauley, J.W. Identification of the glycosaminoglycan-binding site on the glycoprotein Erns of bovine viral diarrhoea virus by site-directed mutagenesis. J. Gen. Virol. 2002, 83, 2153–2159. [Google Scholar] [CrossRef]
- Lussi, C.; de Martin, E.; Schweizer, M. Positively charged amino acids in the pestiviral Erns control cell entry, endoribonuclease activity and innate immune evasion. Viruses 2021, 13, 1581. [Google Scholar] [CrossRef] [PubMed]
- Postel, A.; Smith, D.; Becher, P. Proposed update to the taxonomy of Pestiviruses: Eight additional species within the genus Pestivirus, family Flaviviridae. Viruses 2021, 13, 1542. [Google Scholar] [CrossRef] [PubMed]
- Corapi, W.V.; Donis, O.R.; Dubovi, E.J. Monoclonal antibody analyses of cytopathic and noncytopathic viruses from fatal bovine viral diarrhea virus infections. J. Virol. 1988, 62, 2823–2827. [Google Scholar] [CrossRef] [Green Version]
- Tautz, N.; Meyers, G.; Stark, R.; Dubovi, E.J.; Thiel, H.J. Cytopathogenicity of a pestivirus correlates with a 27-nucleotide insertion. J. Virol. 1996, 70, 7851–7858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkland, P.; Frost, M.; King, K.; Finlaison, D.; Hornitzky, C.; Gu, X.; Richter, M.; Reimann, I.; Dauber, M.; Schirrmeier, H.; et al. Genetic and antigenic characterization of Bungowannah virus, a novel pestivirus. Vet. Microbiol. 2015, 178, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Hause, B.M.; Collin, E.A.; Peddireddi, L.; Yuan, F.; Chen, Z.; Hesse, R.A.; Gauger, P.C.; Clement, T.; Fang, Y.; Anderson, G. Discovery of a novel putative atypical porcine pestivirus in pigs in the USA. J. Gen. Virol. 2015, 96, 2994–2998. [Google Scholar] [CrossRef]
- Stalder, H.; Marti, S.; Flückiger, F.; Renevey, N.; Hofmann, M.A.; Schweizer, M. Complete genome sequences of three border disease virus strains of the same subgenotype, BDSwiss, isolated from sheep, cattle, and pigs in Switzerland. Genome Announc. 2017, 5, e01238-17. [Google Scholar] [CrossRef] [Green Version]
- Postel, A.; Schmeiser, S.; Oğuzoğlu, T.; Indenbirken, D.; Alawi, M.; Fischer, N.; Grundhoff, A.; Becher, P. Close relationship of ruminant pestiviruses and classical swine fever virus. Emerg. Infect. Dis. 2015, 21, 668–672. [Google Scholar] [CrossRef]
- Becher, P.; Fischer, N.; Grundhoff, A.; Stalder, H.; Schweizer, M.; Postel, A. Complete genome sequence of bovine pestivirus strain PG-2, a second member of the tentative pestivirus species giraffe. Genome Announc. 2014, 2, e00376-14. [Google Scholar] [CrossRef] [Green Version]
- Avalos-Ramirez, R.; Orlich, M.; Thiel, H.-J.; Becher, P. Evidence for the presence of two novel pestivirus species. Virology 2001, 286, 456–465. [Google Scholar] [CrossRef] [Green Version]
- Neill, J.D.; Ridpath, J.F.; Fischer, N.; Grundhoff, A.; Postel, A.; Becher, P. Complete genome sequence of pronghorn virus, a pestivirus. Genome Announc. 2014, 2, e00575-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Firth, C.; Bhat, M.; Firth, M.A.; Williams, S.; Frye, M.; Simmonds, P.; Conte, J.M.; Ng, J.; Garcia, J.; Bhuva, N.P.; et al. Detection of zoonotic pathogens and characterization of novel viruses carried by commensal Rattus norvegicus in New York City. mBio 2014, 5, e01933-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Z.; Liu, B.; Du, J.; Zhang, J.; Lu, L.; Zhu, G.; Han, Y.; Su, H.; Yang, L.; Zhang, S.; et al. Discovery of diverse rodent and bat pestiviruses with distinct genomic and phylogenetic characteristics in several Chinese provinces. Front. Microbiol. 2018, 9, 2562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamp, B.; Schwarz, L.; Högler, S.; Riedel, C.; Sinn, L.; Rebel-Bauder, B.; Weissenböck, H.; Ladinig, A.; Rümenapf, T. Novel pestivirus species in pigs, Austria, 2015. Emerg. Infect. Dis. 2017, 23, 1176–1179. [Google Scholar] [CrossRef]
- Jo, W.K.; Van Elk, C.; Van De Bildt, M.; Van Run, P.; Petry, M.; Jesse, S.T.; Jung, K.; Ludlow, M.; Kuiken, T.; Osterhaus, A. An evolutionary divergent pestivirus lacking the Npro gene systemically infects a whale species. Emerg. Microbes Infect. 2019, 8, 1383–1392. [Google Scholar] [CrossRef] [Green Version]
- Gao, W.-H.; Lin, X.-D.; Chen, Y.-M.; Xie, C.-G.; Tan, Z.-Z.; Zhou, J.-J.; Chen, S.; Holmes, E.C.; Zhang, Y.-Z. Newly identified viral genomes in pangolins with fatal disease. Virus Evol. 2020, 6, veaa020. [Google Scholar] [CrossRef]
- Kaiser, V.; Nebel, L.; Schüpbach-Regula, G.; Zanoni, R.G.; Schweizer, M. Influence of border disease virus (BDV) on serological surveillance within the bovine virus diarrhea (BVD) eradication program in Switzerland. BMC Vet. Res. 2017, 13, 21. [Google Scholar] [CrossRef] [Green Version]
- Schweizer, M.; Stalder, H.; Haslebacher, A.; Grisiger, M.; Schwermer, H.; Di Labio, E. Eradication of bovine viral diarrhoea (BVD) in cattle in Switzerland: Lessons taught by the complex biology of the virus. Front. Vet. Sci. 2021, 8, 702730. [Google Scholar] [CrossRef]
- Wegelt, A.; Reimann, I.; Zemke, J.; Beer, M. New insights into processing of bovine viral diarrhea virus glycoproteins Erns and E1. J. Gen. Virol. 2009, 90, 2462–2467. [Google Scholar] [CrossRef]
- Bintintan, I.; Meyers, G. A New type of signal peptidase cleavage site identified in an RNA virus polyprotein. J. Biol. Chem. 2010, 285, 8572–8584. [Google Scholar] [CrossRef] [Green Version]
- Lussi, C.; Sauter, K.-S.; Schweizer, M. Homodimerisation-independent cleavage of dsRNA by a pestiviral nicking endoribonuclease. Sci. Rep. 2018, 8, 8226. [Google Scholar] [CrossRef] [Green Version]
- Backliwal, G.; Hildinger, M.; Kuettel, I.; Delegrange, F.; Hacker, D.L.; Wurm, F.M. Valproic acid: A viable alternative to sodium butyrate for enhancing protein expression in mammalian cell cultures. Biotechnol. Bioeng. 2008, 101, 182–189. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-H.; Chen, L.-H. Promotion of recombinant macrophage colony stimulating factor production by dimethyl sulfoxide addition in Chinese hamster ovary cells. J. Biosci. Bioeng. 2007, 103, 45–49. [Google Scholar] [CrossRef]
- Croset, A.; Delafosse, L.; Gaudry, J.-P.; Arod, C.; Glez, L.; Losberger, C.; Begue, D.; Krstanovic, A.; Robert, F.; Vilbois, F.; et al. Differences in the glycosylation of recombinant proteins expressed in HEK and CHO cells. J. Biotechnol. 2012, 161, 336–348. [Google Scholar] [CrossRef] [PubMed]
- Peterhans, E.; Schweizer, M. BVDV: A pestivirus inducing tolerance of the innate immune response. Biologicals 2013, 41, 39–51. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Mu, Y.; Bintintan, I.; Meyers, G. Downstream sequences control the processing of the pestivirus Erns-E1 precursor. J. Virol. 2020, 95, e01905-20. [Google Scholar] [CrossRef]
- von Heijne, G. The signal peptide. J. Membr. Biol. 1990, 115, 195–201. [Google Scholar] [CrossRef]
- Simmonds, P.; Becher, P.; Collett, M.S.; Gould, E.; Heinz, F.X.; Meyers, G. Flaviviridae. In Virus Taxonomy, 9th ed.; King, A.M.Q., Adams, M.J., Carstens, E.B., Lefkowitz, E.J., Eds.; Academic Press: Oxford, UK, 2011; pp. 1003–1020. [Google Scholar]
- Tao, J.; Li, B.; Chen, J.; Zhang, C.; Ma, Y.; Zhu, G.; Liu, H. Npro His49 and Erns Lys412 mutations in pig bovine viral diarrhea virus type 2 synergistically enhance the cellular antiviral response. Virus Genes 2018, 54, 57–66. [Google Scholar] [CrossRef]
- Karla, A.; Lively, M.O.; Paetzel, M.; Dalbey, R. The identification of residues that control signal peptidase cleavage fidelity and substrate specificity. J. Biol. Chem. 2005, 280, 6731–6741. [Google Scholar] [CrossRef] [Green Version]
- Tews, B.; Klingebeil, A.; Kühn, J.; Franzke, K.; Rümenapf, T.; Meyers, G. The Erns carboxyterminus: Much more than a membrane anchor. Viruses 2021, 13, 1203. [Google Scholar] [CrossRef] [PubMed]
- Krey, T.; Bontems, F.; Vonrhein, C.; Vaney, M.-C.; Bricogne, G.; Rümenapf, T.; Rey, F.A. Crystal structure of the pestivirus envelope glycoprotein Erns and mechanistic analysis of its ribonuclease activity. Structure 2012, 20, 862–873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Becher, P.; Orlich, M.; Shannon, A.D.; Horner, G.; König, M.; Thiel, H.-J. Phylogenetic analysis of pestiviruses from domestic and wild ruminants. J. Gen. Virol. 1997, 78, 1357–1366. [Google Scholar] [CrossRef] [PubMed]
- Harasawa, R.; Giangaspero, M.; Ibata, G.; Paton, D.J. Giraffe strain of pestivirus: Its taxonomic status based on the 5′-untranslated region. Microbiol. Immunol. 2000, 44, 915–921. [Google Scholar] [CrossRef]
- Schmeiser, S.; Mast, J.; Thiel, H.-J.; König, M.; Sandri-Goldin, R.M. Morphogenesis of pestiviruses: New insights from ultrastructural studies of strain Giraffe-1. J. Virol. 2014, 88, 2717–2724. [Google Scholar] [CrossRef] [Green Version]
- Gutekunst, E.D.; Malmquist, A.W. Separation of a soluble antigen and infectious particles of bovine viral diarrhea viruses and their relationship to hog cholera. Can. J. Comp. Med. Vet. Sci. 1963, 27, 121–123. [Google Scholar]
- Thiel, H.J.; Stark, R.; Weiland, E.; Rümenapf, T.; Meyers, G. Hog cholera virus: Molecular composition of virions from a pestivirus. J. Virol. 1991, 65, 4705–4712. [Google Scholar] [CrossRef] [Green Version]
- Tews, B.A.; Schürmann, E.-M.; Meyers, G. Mutation of cysteine 171 of pestivirus Erns RNase prevents homodimer formation and leads to attenuation of classical swine fever virus. J. Virol. 2009, 83, 4823–4834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van Gennip, H.G.P.; Hesselink, A.T.; Moormann, R.J.M.; Hulst, M.M. Dimerisation of glycoprotein Erns of classical swine fever virus is not essential for viral replication and infection. Arch. Virol. 2005, 150, 2271–2286. [Google Scholar] [CrossRef] [PubMed]
- Riedel, C.; Aitkenhead, H.; El Omari, K.; Rümenapf, T. Atypical porcine pestiviruses: Relationships and conserved structural features. Viruses 2021, 13, 760. [Google Scholar] [CrossRef] [PubMed]
- Szillat, K.P.; Koethe, S.; Wernike, K.; Höper, D.; Beer, M. A CRISPR/Cas9 generated bovine CD46-knockout cell line—A tool to elucidate the adaptability of bovine viral diarrhea viruses (BVDV). Viruses 2020, 12, 859. [Google Scholar] [CrossRef]

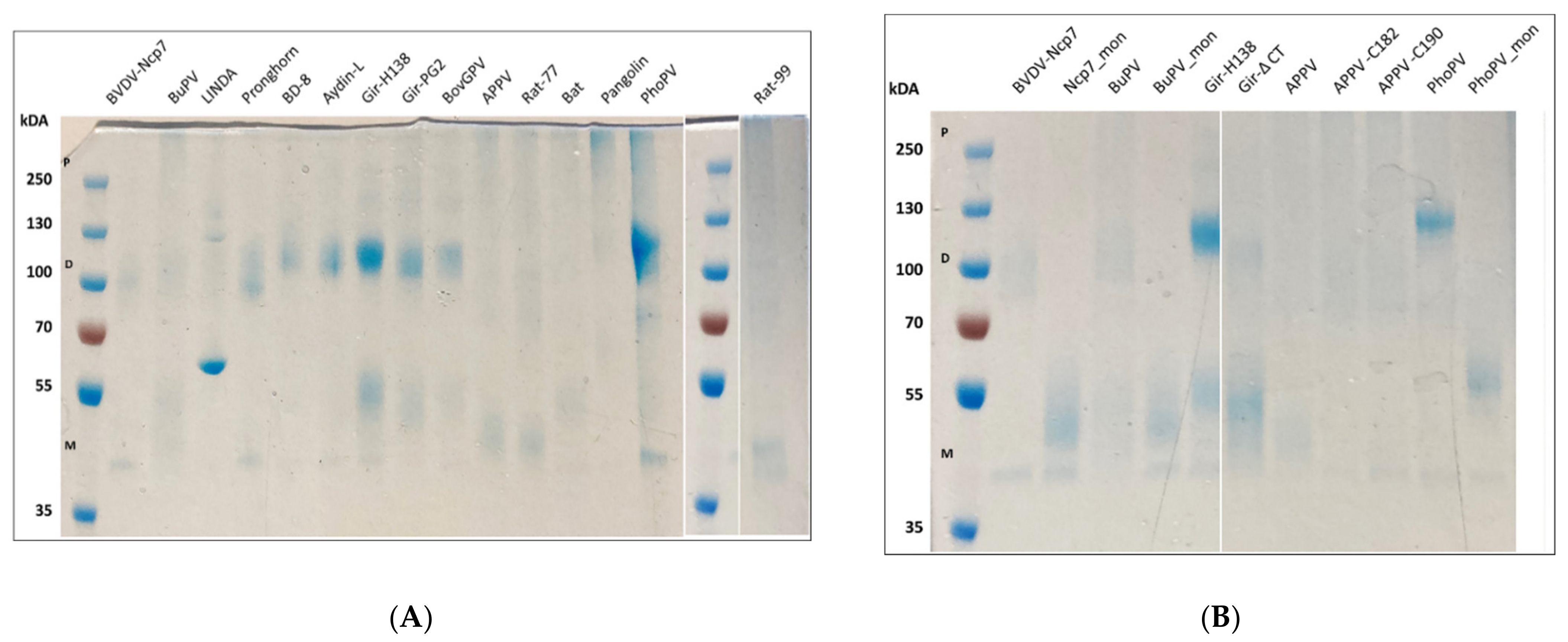
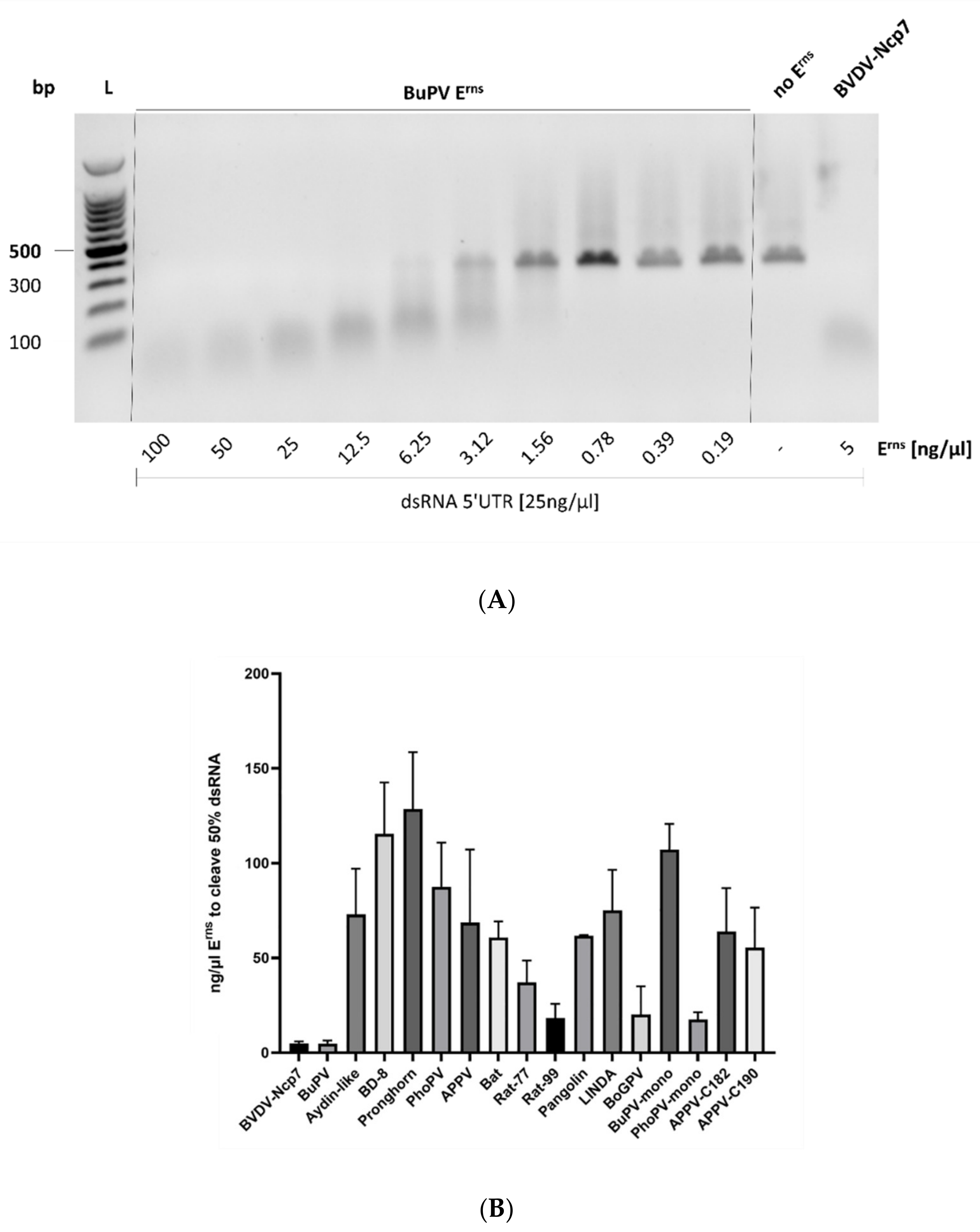
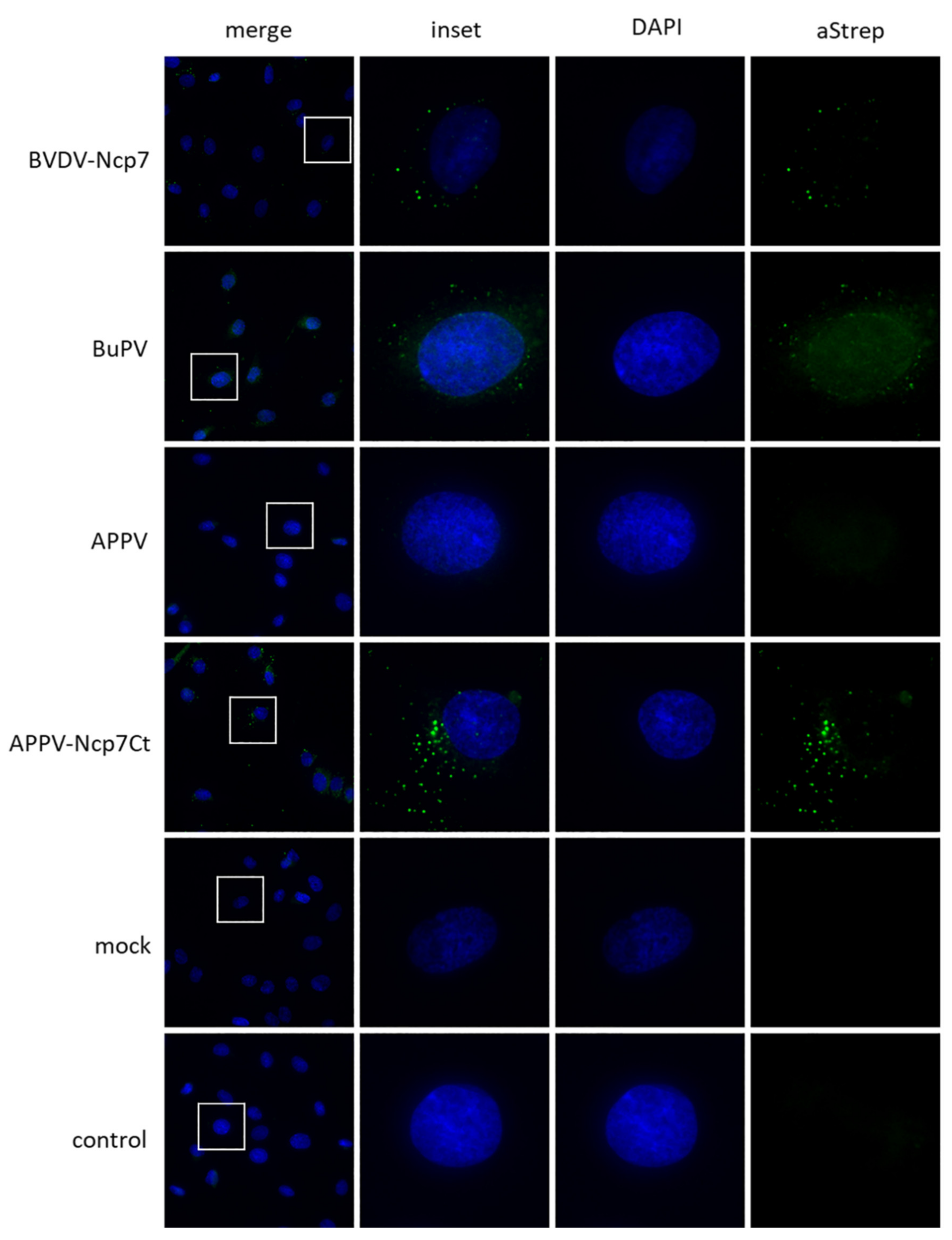
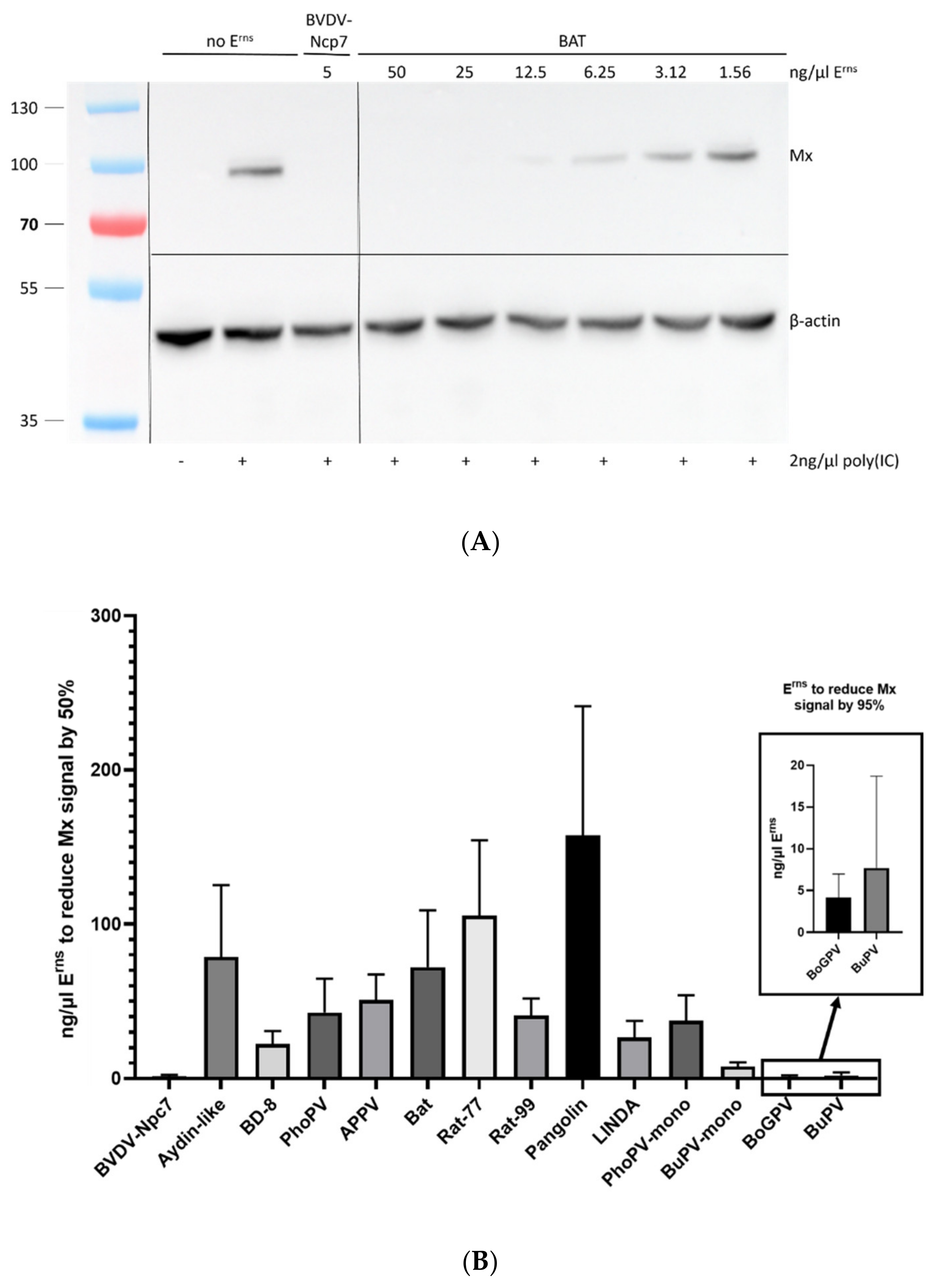
| Nomenclature Used | GenBank no. | Species | Reference | RNase Assay [%] | Mx Assay [%] |
|---|---|---|---|---|---|
| BVDV Ncp7 | n.a. | A | [26,27] | =100 | =100 |
| BuPV (Bungowannah) | NC_023176 | F | [28] | 86.24 | 387.5 |
| APPV | KR011347 | K | [29] | 6.11 | 6.07 |
| BD8 (Border Disease) | R4785/06 | D | [30] | 3.63 | 13.82 |
| Aydin-like | JX428945 | I | [31] | 5.75 | 3.93 |
| Gir-PG2 (Giraffe) | KJ660072 | G | [32] | <2.1 (3) | <1.55 (3) |
| Gir-H138 (Giraffe) | AF144617 | G | [33] | <2.1 (3) | <1.55 (3) |
| Pronghorn | AY781152 | E | [34] | 3.26 | <1.55 (3) |
| Rat-77 | NC_025677 | J | [35] | 11.31 | 2.93 |
| Rat-99 | KY370099 | Q (1) | [36] | 22.92 | 7.59 |
| LINDA | KY436034 | L (1) | [37] | 5.58 | 11.62 |
| PhoPV (phocoena) | NS170385 | M (1) | [38] | 4.79 | 7.25 |
| Pangolin | MK636875 | P (1) | [39] | 6.80 | 1.97 |
| Bat | MH282908 | S (1) | [36] | 6.90 | 4.29 |
| BoGPV (Giraffe) | n.a. | G | this paper (2) | 20.76 | 688.89 |
| BuPV-mono | n.a. | F | this paper | 3.91 | 39.34 |
| PhoPV-mono | n.a. | M (1) | this paper | 23.91 | 8.22 |
| APPV-C182 | n.a. | K | this paper | 6.56 | n.d. |
| APPV-C190 | n.a. | K | this paper | 7.58 | n.d. |
| H138-Ncp7-1 | n.a. | chimera | this paper | 2.63 | n.d. |
| H138-Ncp7-2 | n.a. | chimera | this paper | <2.1 (3) | n.d. |
| Giraffe ΔC | n.a. | G | this paper | <2.1 (3) | n.d. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
de Martin, E.; Schweizer, M. Fifty Shades of Erns: Innate Immune Evasion by the Viral Endonucleases of All Pestivirus Species. Viruses 2022, 14, 265. https://doi.org/10.3390/v14020265
de Martin E, Schweizer M. Fifty Shades of Erns: Innate Immune Evasion by the Viral Endonucleases of All Pestivirus Species. Viruses. 2022; 14(2):265. https://doi.org/10.3390/v14020265
Chicago/Turabian Stylede Martin, Elena, and Matthias Schweizer. 2022. "Fifty Shades of Erns: Innate Immune Evasion by the Viral Endonucleases of All Pestivirus Species" Viruses 14, no. 2: 265. https://doi.org/10.3390/v14020265






