StREM1.3 REMORIN Protein Plays an Agonistic Role in Potyvirus Cell-to-Cell Movement in N. benthamiana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Molecular Clones and Fluorescent Tagged Potyvirus Infectious Clones
2.3. GUS Assays
2.4. Transient Protein Expression and Confocal Microscopy Analyses
2.5. Callose Quantification by Aniline Blue Staining and Colocalisation Analysis
2.6. RNA Extraction and RT–qPCR
2.7. Interaction Analyses through Split-Ubiquitin Y2H and BiFC Assays
2.8. Protein Analysis, Western Blots
2.9. Statistical Analysis
3. Results
3.1. StREM1.3 Overexpression Increases Potyvirus Propagation in N. benthamiana
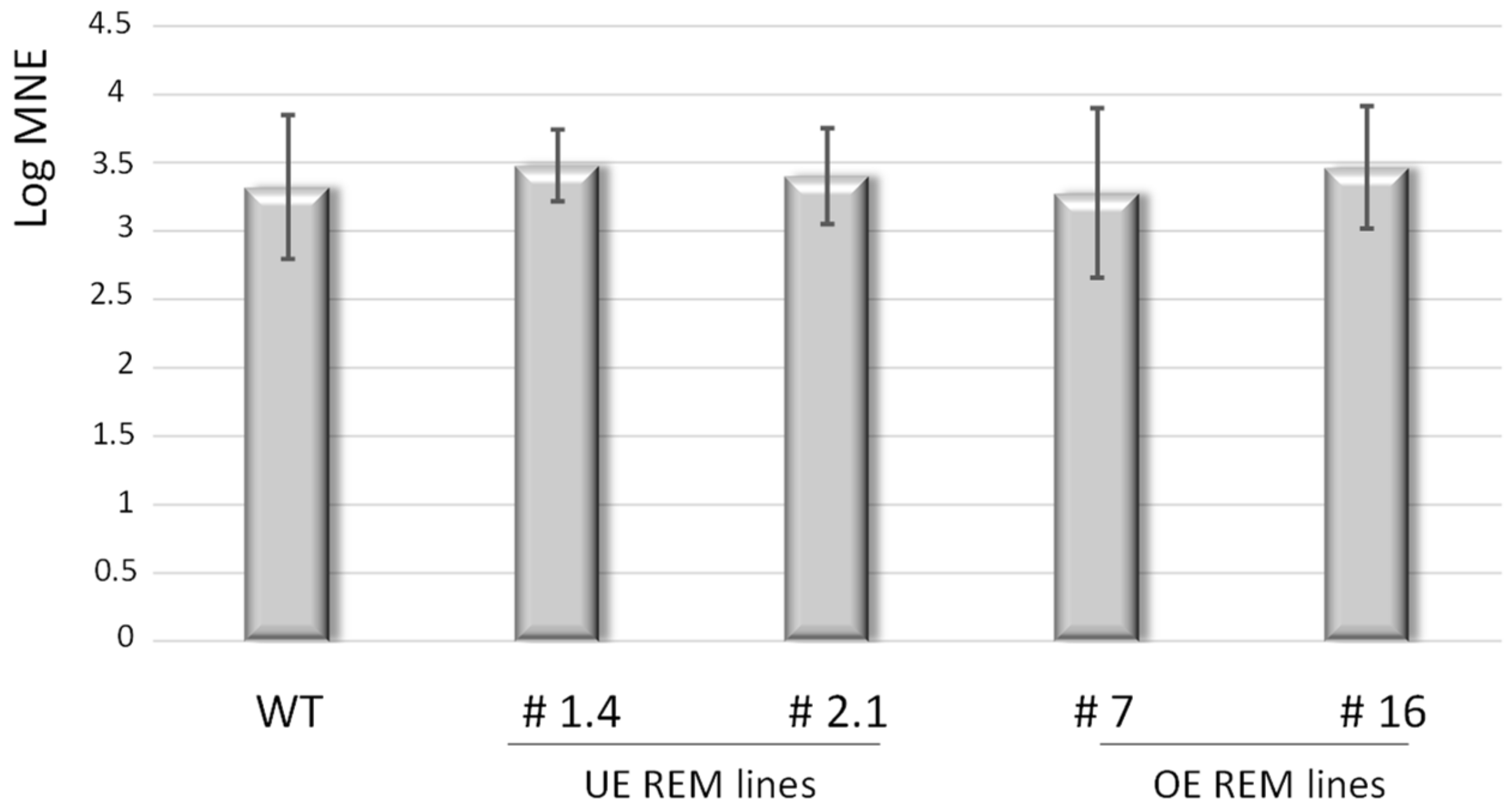
3.2. The Propagation of TuMV Is Slowed Down in Lines Underexpressing Group 1 Endogenous NbREMs
3.3. StREM1.3 and NbREM Expression Levels Do Not Affect TuMV Replication
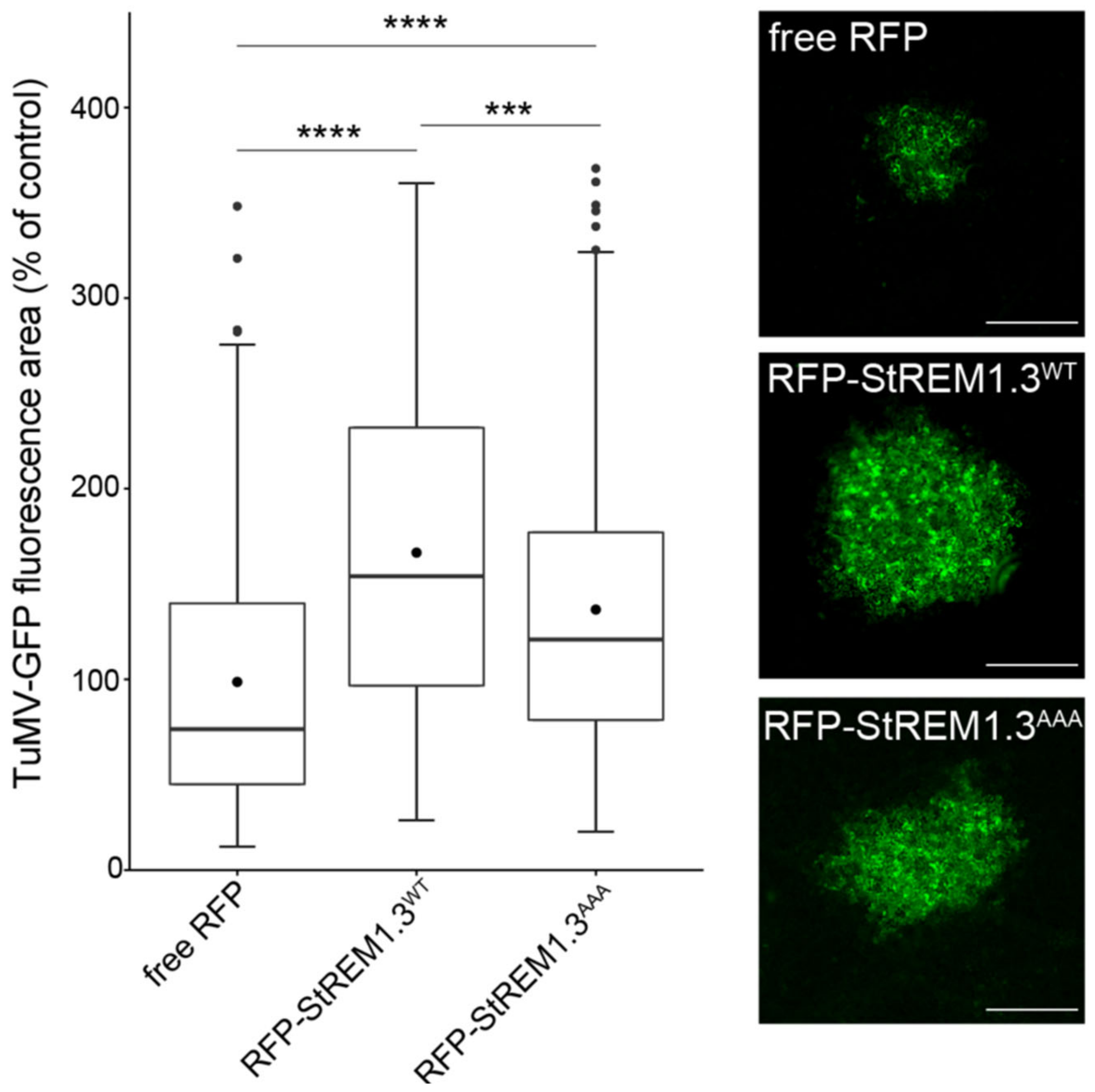
3.4. StREM1.3 Membrane Anchoring Is Necessary for Increasing Potyvirus Cell-to-Cell Movement
3.5. StREM1.3 Phosphorylation Is Involved in Increasing Potyvirus Cell-to-Cell Movement
3.6. TuMV Infection Does Not Alter the Expression Level of Endogenous NbREM, Nor the Membranous Localisation of GFP–StREM1.3
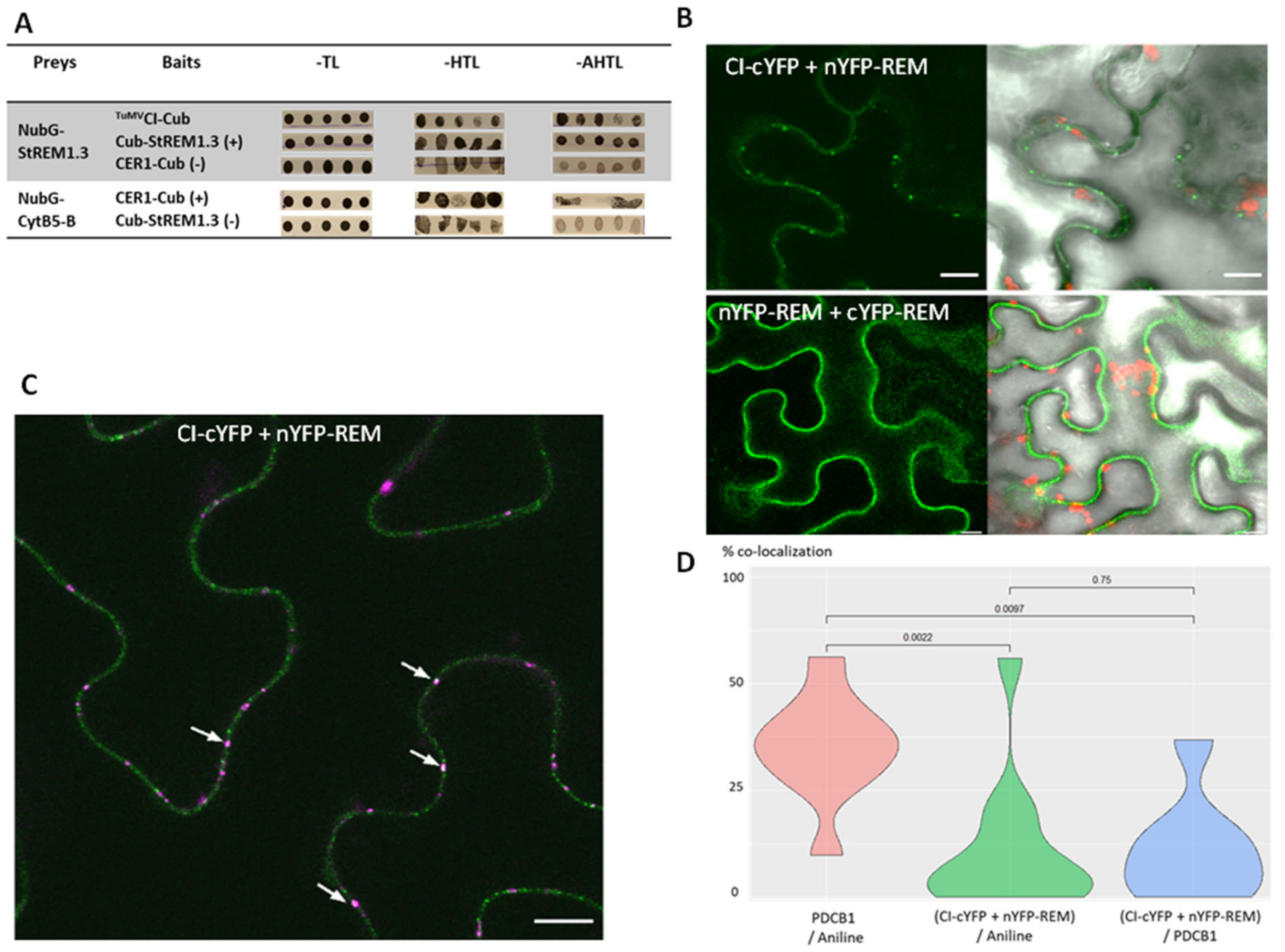
3.7. StREM1.3 Interacts in Yeast and in Planta with TuMVCI, a Viral Protein Crucial for Potyvirus Movement
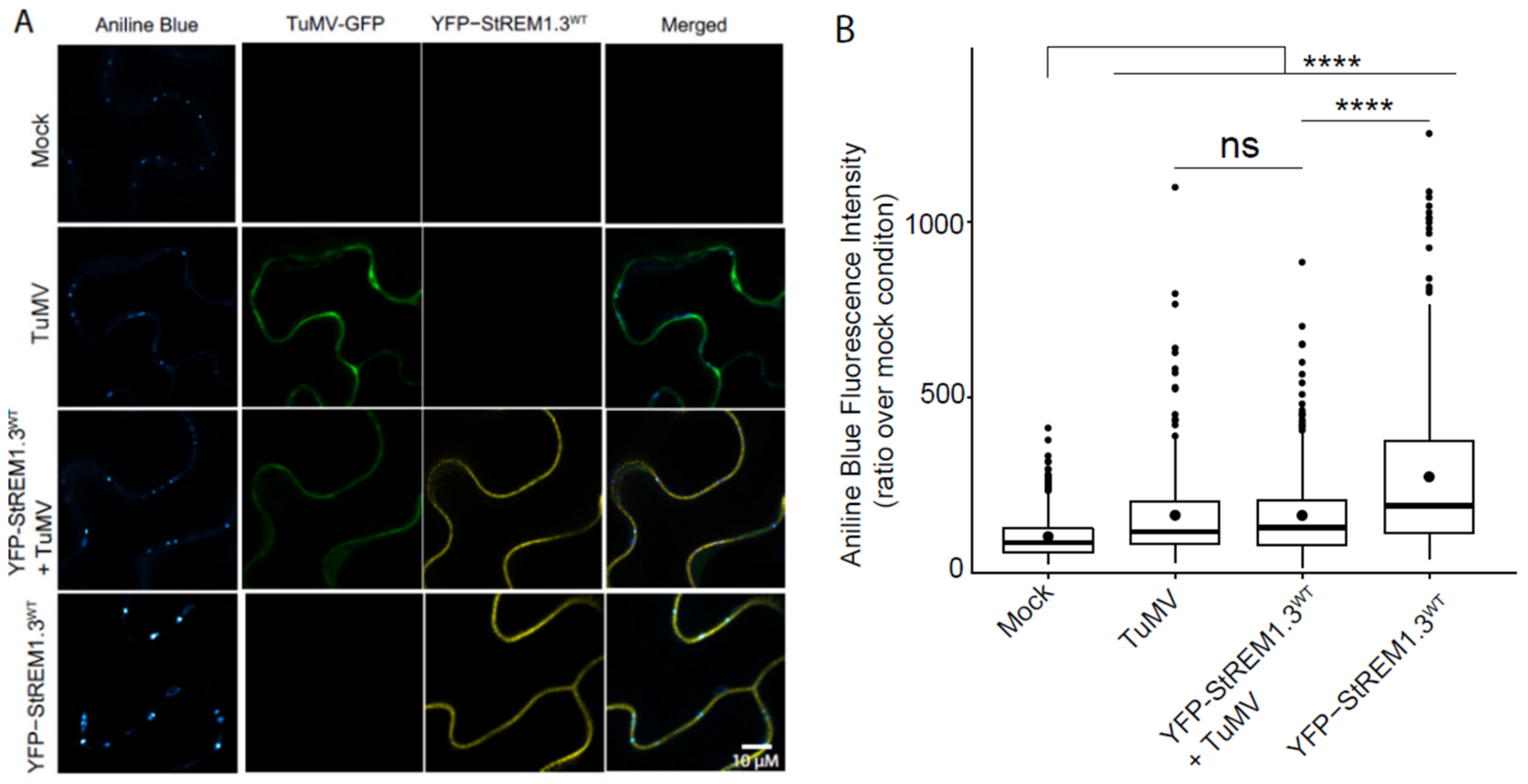
3.8. Callose Deposition Induced by StREM1.3 Is Decreased by TuMV Infection
4. Discussion
4.1. REMORIN Proteins Have Antagonistic or Agonistic Roles in Viral Cell-to-Cell Movement
4.2. TuMV Infection in N. benthamiana Does Not Affect Neither the Levels of Endogenous REMORIN Proteins, Nor the Plasma Membrane Localisation of Fluorescently Tagged StREM1.3
4.3. TuMV Infection Interferes with the StREM1.3-Mediated Callose Deposition in N. benthamiana
4.4. The Interaction between TuMVCI and StREM1.3 Occurs at the PM but Is Not Predominantly Associated to PDs: Could This Interaction Play a Role in the Non-PD Movement of Potyviruses?
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Reagan, B.C.; Burch-Smith, T.M. Viruses Reveal the Secrets of Plasmodesmal Cell Biology. Mol. Plant-Microbe Interact. 2020, 33, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Schoelz, J.E.; Harries, P.A.; Nelson, R.S. Intracellular Transport of Plant Viruses: Finding the Door out of the Cell. Mol. Plant 2011, 4, 813–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heinlein, M. Plasmodesmata: Channels for Viruses on the Move. In Plasmodesmata; Heinlein, M., Ed.; Springer: New York, NY, USA, 2015; Volume 1217, pp. 25–52. ISBN 978-1-4939-1522-4. [Google Scholar]
- Wylie, S.J.; Adams, M.; Chalam, C.; Kreuze, J.; López-Moya, J.J.; Ohshima, K.; Praveen, S.; Rabenstein, F.; Stenger, D.; Wang, A.; et al. ICTV Virus Taxonomy Profile: Potyviridae. J. Gen. Virol. 2017, 98, 352–354. [Google Scholar] [CrossRef] [PubMed]
- Wang, A. Cell-to-Cell Movement of Plant Viruses via Plasmodesmata: A Current Perspective on Potyviruses. Curr. Opin. Virol. 2021, 48, 10–16. [Google Scholar] [CrossRef]
- Deng, P.; Wu, Z.; Wang, A. The Multifunctional Protein CI of Potyviruses Plays Interlinked and Distinct Roles in Viral Genome Replication and Intercellular Movement. Virol. J. 2015, 12, 141. [Google Scholar] [CrossRef] [Green Version]
- Movahed, N.; Patarroyo, C.; Sun, J.; Vali, H.; Laliberté, J.-F.; Zheng, H. Cylindrical Inclusion Protein of Turnip Mosaic Virus Serves as a Docking Point for the Intercellular Movement of Viral Replication Vesicles. Plant Physiol. 2017, 175, 1732–1744. [Google Scholar] [CrossRef] [Green Version]
- Sorel, M.; Garcia, J.A.; German-Retana, S. The Potyviridae Cylindrical Inclusion Helicase: A Key Multipartner and Multifunctional Protein. Mol. Plant-Microbe Interact. 2014, 27, 215–226. [Google Scholar] [CrossRef] [Green Version]
- Wei, T.; Zhang, C.; Hong, J.; Xiong, R.; Kasschau, K.D.; Zhou, X.; Carrington, J.C.; Wang, A. Formation of Complexes at Plasmodesmata for Potyvirus Intercellular Movement Is Mediated by the Viral Protein P3N-PIPO. PLoS Pathog. 2010, 6, e1000962. [Google Scholar] [CrossRef] [Green Version]
- Beauchemin, C.; Boutet, N.; Laliberte, J.F. Visualization of the Interaction between the Precursors of VPg, the Viral Protein Linked to the Genome of Turnip Mosaic Virus, and the Translation Eukaryotic Initiation Factor Iso 4E in Planta. J. Virol. 2007, 81, 775–782. [Google Scholar] [CrossRef] [Green Version]
- Cotton, S.; Grangeon, R.; Thivierge, K.; Mathieu, I.; Ide, C.; Wei, T.; Wang, A.; Laliberté, J.-F. Turnip Mosaic Virus RNA Replication Complex Vesicles Are Mobile, Align with Microfilaments, and Are Each Derived from a Single Viral Genome. J. Virol. 2009, 83, 10460–10471. [Google Scholar] [CrossRef] [Green Version]
- Grangeon, R.; Jiang, J.; Wan, J.; Agbeci, M.; Zheng, H.; Laliberté, J.-F. 6K2-Induced Vesicles Can Move Cell to Cell during Turnip Mosaic Virus Infection. Front. Microbiol. 2013, 4, 351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.; Patarroyo, C.; Garcia Cabanillas, D.; Zheng, H.; Laliberté, J.-F. The Vesicle-Forming 6K 2 Protein of Turnip Mosaic Virus Interacts with the COPII Coatomer Sec24a for Viral Systemic Infection. J. Virol. 2015, 89, 6695–6710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaad, M.C.; Jensen, P.E.; Carrington, J.C. Formation of Plant RNA Virus Replication Complexes on Membranes: Role of an Endoplasmic Reticulum-Targeted Viral Protein. EMBO J. 1997, 16, 4049–4059. [Google Scholar] [CrossRef] [PubMed]
- Chai, M.; Wu, X.; Liu, J.; Fang, Y.; Luan, Y.; Cui, X.; Zhou, X.; Wang, A.; Cheng, X. P3N-PIPO Interacts with P3 via the Shared N-Terminal Domain To Recruit Viral Replication Vesicles for Cell-to-Cell Movement. J. Virol. 2020, 94, e01898-19. [Google Scholar] [CrossRef] [PubMed]
- Den Boon, J.A.; Diaz, A.; Ahlquist, P. Cytoplasmic Viral Replication Complexes. Cell Host Microbe 2010, 8, 77–85. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cabanillas, D.G.; Jiang, J.; Movahed, N.; Germain, H.; Yamaji, Y.; Zheng, H.; Laliberté, J.-F. Turnip Mosaic Virus Uses the SNARE Protein VTI11 in an Unconventional Route for Replication Vesicle Trafficking. Plant Cell 2018, 30, 2594–2615. [Google Scholar] [CrossRef] [Green Version]
- Movahed, N.; Cabanillas, D.G.; Wan, J.; Vali, H.; Laliberté, J.-F.; Zheng, H. Turnip Mosaic Virus Components Are Released into the Extracellular Space by Vesicles in Infected Leaves. Plant Physiol. 2019, 180, 1375–1388. [Google Scholar] [CrossRef]
- Vijayapalani, P.; Maeshima, M.; Nagasaki-Takekuchi, N.; Miller, W.A. Interaction of the Trans-Frame Potyvirus Protein P3N-PIPO with Host Protein PCaP1 Facilitates Potyvirus Movement. PLoS Pathog. 2012, 8, e1002639. [Google Scholar] [CrossRef]
- Park, S.-H.; Li, F.; Renaud, J.; Shen, W.; Li, Y.; Guo, L.; Cui, H.; Sumarah, M.; Wang, A. NbEXPA1, an α-Expansin, Is Plasmodesmata-Specific and a Novel Host Factor for Potyviral Infection. Plant J. 2017, 92, 846–861. [Google Scholar] [CrossRef] [Green Version]
- Hofius, D.; Maier, A.T.; Dietrich, C.; Jungkunz, I.; Bornke, F.; Maiss, E.; Sonnewald, U. Capsid Protein-Mediated Recruitment of Host DnaJ-Like Proteins Is Required for Potato Virus Y Infection in Tobacco Plants. J. Virol. 2007, 81, 11870–11880. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Lu, L.; Wang, Y.; Yuan, X.; Chen, X. The Interaction of Soybean Reticulon Homology Domain Protein (GmRHP) with Soybean Mosaic Virus Encoded P3 Contributes to the Viral Infection. Biochem. Biophys. Res. Commun. 2018, 495, 2105–2110. [Google Scholar] [CrossRef]
- Geng, C.; Cong, Q.-Q.; Li, X.-D.; Mou, A.-L.; Gao, R.; Liu, J.-L.; Tian, Y.-P. Developmentally regulated plasma membrane protein of Nicotiana benthamiana Contributes to Potyvirus Movement and Transports to Plasmodesmata via the Early Secretory Pathway and the Actomyosin System. Plant Physiol. 2015, 167, 394–410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Movahed, N.; Sun, J.; Vali, H.; Laliberté, J.-F.; Zheng, H. A Host ER Fusogen Is Recruited by Turnip Mosaic Virus for Maturation of Viral Replication Vesicles. Plant Physiol. 2019, 179, 507–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Uchiyama, A.; Shimada-Beltran, H.; Levy, A.; Zheng, J.Y.; Javia, P.A.; Lazarowitz, S.G. The Arabidopsis Synaptotagmin SYTA Regulates the Cell-to-Cell Movement of Diverse Plant Viruses. Front. Plant Sci. 2014, 5, 584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, G.; Cui, X.; Chen, H.; Renaud, J.B.; Yu, K.; Chen, X.; Wang, A. Dynamin-Like Proteins of Endocytosis in Plants Are Coopted by Potyviruses To Enhance Virus Infection. J. Virol. 2018, 92, e01320-18. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Cui, X.; Dai, Z.; He, R.; Li, Y.; Yu, K.; Bernards, M.; Chen, X.; Wang, A. A Plant RNA Virus Hijacks Endocytic Proteins to Establish Its Infection in Plants. Plant J. 2020, 101, 384–400. [Google Scholar] [CrossRef]
- Wu, G.; Jia, Z.; Ding, K.; Zheng, H.; Lu, Y.; Lin, L.; Peng, J.; Rao, S.; Wang, A.; Chen, J.; et al. Turnip Mosaic Virus Co-Opts the Vacuolar Sorting Receptor VSR4 to Promote Viral Genome Replication in Plants by Targeting Viral Replication Vesicles to the Endosome. PLoS Pathog. 2022, 18, e1010257. [Google Scholar] [CrossRef]
- Cheng, G.; Yang, Z.; Zhang, H.; Zhang, J.; Xu, J. Remorin Interacting with PCaP1 Impairs Turnip Mosaic Virus Intercellular Movement but Is Antagonised by VPg. New Phytol. 2020, 225, 2122–2139. [Google Scholar] [CrossRef]
- Gouguet, P.; Gronnier, J.; Legrand, A.; Perraki, A.; Jolivet, M.-D.; Deroubaix, A.-F.; German-Retana, S.; Boudsocq, M.; Habenstein, B.; Mongrand, S.; et al. Connecting the Dots: From Nanodomains to Physiological Functions of REMORINs. Plant Physiol. 2020, 185, 632–649. [Google Scholar] [CrossRef]
- Raffaele, S.; Mongrand, S.; Gamas, P.; Niebel, A.; Ott, T. Genome-Wide Annotation of Remorins, a Plant-Specific Protein Family: Evolutionary and Functional Perspectives. Plant Physiol. 2007, 145, 593–600. [Google Scholar] [CrossRef] [Green Version]
- Perraki, A.; Gronnier, J.; Gouguet, P.; Boudsocq, M.; Deroubaix, A.-F.; Simon, V.; German-Retana, S.; Legrand, A.; Habenstein, B.; Zipfel, C.; et al. REM1.3’s Phospho-Status Defines Its Plasma Membrane Nanodomain Organization and Activity in Restricting PVX Cell-to-Cell Movement. PLoS Pathog. 2018, 14, e1007378. [Google Scholar] [CrossRef] [PubMed]
- Raffaele, S.; Bayer, E.; Mongrand, S. Upregulation of the Plant Protein Remorin Correlates with Dehiscence and Cell Maturation: A Link with the Maturation of Plasmodesmata? Plant Signal. Behav. 2009, 4, 915–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marín, M.; Ott, T. Phosphorylation of Intrinsically Disordered Regions in Remorin Proteins. Front. Plant Sci. 2012, 3, 86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gronnier, J.; Crowet, J.-M.; Habenstein, B.; Nasir, M.N.; Bayle, V.; Hosy, E.; Platre, M.P.; Gouguet, P.; Raffaele, S.; Martinez, D.; et al. Structural Basis for Plant Plasma Membrane Protein Dynamics and Organization into Functional Nanodomains. eLife 2017, 6, e26404. [Google Scholar] [CrossRef] [PubMed]
- Perraki, A.; Cacas, J.L.; Crowet, J.M.; Lins, L.; Castroviejo, M.; German-Retana, S.; Mongrand, S.; Raffaele, S. Plasma Membrane Localization of Solanum Tuberosum Remorin from Group 1, Homolog 3 Is Mediated by Conformational Changes in a Novel C-Terminal Anchor and Required for the Restriction of Potato Virus X Movement. Plant Physiol. 2012, 160, 624–637. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.M.; Chng, C.-P.; Pu, X.; Ma, Z.; Han, X.; Liu, X.; Yang, L.; Huang, C.; Miao, Y. Potentiation of Plant Defense by Bacterial Outer Membrane Vesicles Is Mediated by Membrane Nanodomains. Plant Cell 2021, 34, 395–417. [Google Scholar] [CrossRef]
- Rutter, B.D.; Innes, R.W. Extracellular Vesicles Isolated from the Leaf Apoplast Carry Stress-Response Proteins. Plant Physiol. 2017, 173, 728–741. [Google Scholar] [CrossRef] [Green Version]
- Rutter, B.D.; Innes, R.W. Extracellular Vesicles as Key Mediators of Plant–Microbe Interactions. Curr. Opin. Plant Biol. 2018, 44, 16–22. [Google Scholar] [CrossRef]
- Raffaele, S.; Bayer, E.; Lafarge, D.; Cluzet, S.; German Retana, S.; Boubekeur, T.; Leborgne-Castel, N.; Carde, J.-P.; Lherminier, J.; Noirot, E.; et al. Remorin, a Solanaceae Protein Resident in Membrane Rafts and Plasmodesmata, Impairs Potato Virus X Movement. Plant Cell 2009, 21, 1541–1555. [Google Scholar] [CrossRef] [Green Version]
- Son, S.; Oh, C.J.; An, C.S. Arabidopsis Thaliana Remorins Interact with SnRK1 and Play a Role in Susceptibility to Beet Curly Top Virus and Beet Severe Curly Top Virus. Plant Pathol. J. 2014, 30, 269–278. [Google Scholar] [CrossRef] [Green Version]
- Fu, S.; Xu, Y.; Li, C.; Li, Y.; Wu, J.; Zhou, X. Rice Stripe Virus Interferes with S-Acylation of Remorin and Induces Its Autophagic Degradation to Facilitate Virus Infection. Mol. Plant 2018, 11, 269–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brault, M.L.; Petit, J.D.; Immel, F.; Nicolas, W.J.; Glavier, M.; Brocard, L.; Gaston, A.; Fouché, M.; Hawkins, T.J.; Crowet, J.; et al. Multiple C2 Domains and Transmembrane Region Proteins (MCTP s) Tether Membranes at Plasmodesmata. EMBO Rep. 2019, 20, e47182. [Google Scholar] [CrossRef] [PubMed]
- Beauchemin, C.; Bougie, V.; Laliberte, J.F. Simultaneous Production of Two Foreign Proteins from a Polyvirus-Based Vector. Virus Res. 2005, 112, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, K.I.; Puustinen, P.; Gabrenaite, R.; Vihinen, H.; Ronnstrand, L.; Valmu, L.; Kalkkinen, N.; Makinen, K. Phosphorylation of the Potyvirus Capsid Protein by Protein Kinase CK2 and Its Relevance for Virus Infection. Plant Cell 2003, 15, 2124–2139. [Google Scholar] [CrossRef] [Green Version]
- German-Retana, S.; Candresse, T.; Alias, E.; Delbos, R.; Le Gall, O. Effects of GFP or GUS Tagging on the Accumulation and Pathogenicity of a Resistance Breaking LMV Isolate in Susceptible and Resistant Lettuce Cultivars. Mol. Plant-Microbe Interact. 2000, 13, 316–324. [Google Scholar] [CrossRef] [Green Version]
- Ito, Y.; Esnay, N.; Platre, M.P.; Wattelet-Boyer, V.; Noack, L.C.; Fougère, L.; Menzel, W.; Claverol, S.; Fouillen, L.; Moreau, P.; et al. Sphingolipids Mediate Polar Sorting of PIN2 through Phosphoinositide Consumption at the Trans-Golgi Network. Nat. Commun. 2021, 12, 4267. [Google Scholar] [CrossRef]
- Simon, P. Q-Gene: Processing Quantitative Real-Time RT-PCR Data. Bioinformatics 2003, 19, 1439–1440. [Google Scholar] [CrossRef] [Green Version]
- Bernard, A.; Domergue, F.; Pascal, S.; Jetter, R.; Renne, C.; Faure, J.-D.; Haslam, R.P.; Napier, J.A.; Lessire, R.; Joubes, J. Reconstitution of Plant Alkane Biosynthesis in Yeast Demonstrates That Arabidopsis ECERIFERUM1 and ECERIFERUM3 Are Core Components of a Very-Long-Chain Alkane Synthesis Complex. Plant Cell 2012, 24, 3106–3118. [Google Scholar] [CrossRef] [Green Version]
- Martin, K.; Kopperud, K.; Chakrabarty, R.; Banerjee, R.; Brooks, R.; Goodin, M.M. Transient Expression in Nicotiana benthamiana Fluorescent Marker Lines Provides Enhanced Definition of Protein Localization, Movement and Interactions in Planta. Plant J. 2009, 59, 150–162. [Google Scholar] [CrossRef]
- Legrand, A.; Martinez, D.; Grélard, A.; Berbon, M.; Morvan, E.; Tawani, A.; Loquet, A.; Mongrand, S.; Habenstein, B. Nanodomain Clustering of the Plant Protein Remorin by Solid-State NMR. Front. Mol. Biosci. 2019, 6, 107. [Google Scholar] [CrossRef] [Green Version]
- Martinez, D.; Legrand, A.; Gronnier, J.; Decossas, M.; Gouguet, P.; Lambert, O.; Berbon, M.; Verron, L.; Grélard, A.; Germain, V.; et al. Coiled-Coil Oligomerization Controls Localization of the Plasma Membrane REMORINs. J. Struct. Biol. 2019, 206, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Roberts, I.M.; Wang, D.; Findlay, K.; Maule, A.J. Ultrastructural and Temporal Observations of the Potyvirus Cylindrical Inclusions (Cls) Show That the Cl Protein Acts Transiently in Aiding Virus Movement. Virology 1998, 245, 173–181. [Google Scholar] [CrossRef] [Green Version]
- Sasaki, N.; Takashima, E.; Nyunoya, H. Altered Subcellular Localization of a Tobacco Membrane Raft-Associated Remorin Protein by Tobamovirus Infection and Transient Expression of Viral Replication and Movement Proteins. Front. Plant Sci. 2018, 9, 619. [Google Scholar] [CrossRef] [PubMed]
- Perraki, A.; Binaghi, M.; Mecchia, M.A.; Gronnier, J.; German-Retana, S.; Mongrand, S.; Bayer, E.; Zelada, A.M.; Germain, V. StRemorin1.3 Hampers Potato Virus X TGBp1 Ability to Increase Plasmodesmata Permeability, but Does Not Interfere with Its Silencing Suppressor Activity. FEBS Lett. 2014, 588, 1699–1705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Storme, N.; Geelen, D. Callose Homeostasis at Plasmodesmata: Molecular Regulators and Developmental Relevance. Front. Plant Sci. 2014, 5, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zavaliev, R.; Levy, A.; Gera, A.; Epel, B.L. Subcellular Dynamics and Role of Arabidopsis β-1,3-Glucanases in Cell-to-Cell Movement of Tobamoviruses. Mol. Plant-Microbe Interact. 2013, 26, 1016–1030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, J.; Laliberté, J.-F. Membrane-Associated Virus Replication Complexes Locate to Plant Conducting Tubes. Plant Signal. Behav. 2015, 10, e1042639. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| Primer Name | Sequence (5′–3′) |
|---|---|
| CP1-F | TCAAACCATGTGTCAAACTCG |
| CP2-R | CGAGAAAAGAGTGGCCCTAA |
| GFP-HDEL_F | CCTGTCCTTTTACCAGAC |
| GFP-HDEL_R | CTCGTCATGAGATCTGTATAG |
| TuMVCI-SfiI-F | ATTAACAAGGCCATTACGGCCATGACTCTCAATGATATAGAGGA |
| TuMVCI-SfiI-R | AACTGATTGGCCGAGGCGGCCCCCTATTGATGGTGAACTGCCTC |
| St-REM1.3-SfiI-F | ATTAACAAGGCCATTACGGCCATGGCAGAATTGGAAGCTAAGAAAG |
| St-REM1.3-SfiI-R | AACTGATTGGCCGAGGCGGCCTCAAAATATTCCAAGGATTTTCTTTGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rocher, M.; Simon, V.; Jolivet, M.-D.; Sofer, L.; Deroubaix, A.-F.; Germain, V.; Mongrand, S.; German-Retana, S. StREM1.3 REMORIN Protein Plays an Agonistic Role in Potyvirus Cell-to-Cell Movement in N. benthamiana. Viruses 2022, 14, 574. https://doi.org/10.3390/v14030574
Rocher M, Simon V, Jolivet M-D, Sofer L, Deroubaix A-F, Germain V, Mongrand S, German-Retana S. StREM1.3 REMORIN Protein Plays an Agonistic Role in Potyvirus Cell-to-Cell Movement in N. benthamiana. Viruses. 2022; 14(3):574. https://doi.org/10.3390/v14030574
Chicago/Turabian StyleRocher, Marion, Vincent Simon, Marie-Dominique Jolivet, Luc Sofer, Anne-Flore Deroubaix, Véronique Germain, Sébastien Mongrand, and Sylvie German-Retana. 2022. "StREM1.3 REMORIN Protein Plays an Agonistic Role in Potyvirus Cell-to-Cell Movement in N. benthamiana" Viruses 14, no. 3: 574. https://doi.org/10.3390/v14030574






