A Review of Pseudorabies Virus Variants: Genomics, Vaccination, Transmission, and Zoonotic Potential
Abstract
:1. Introduction
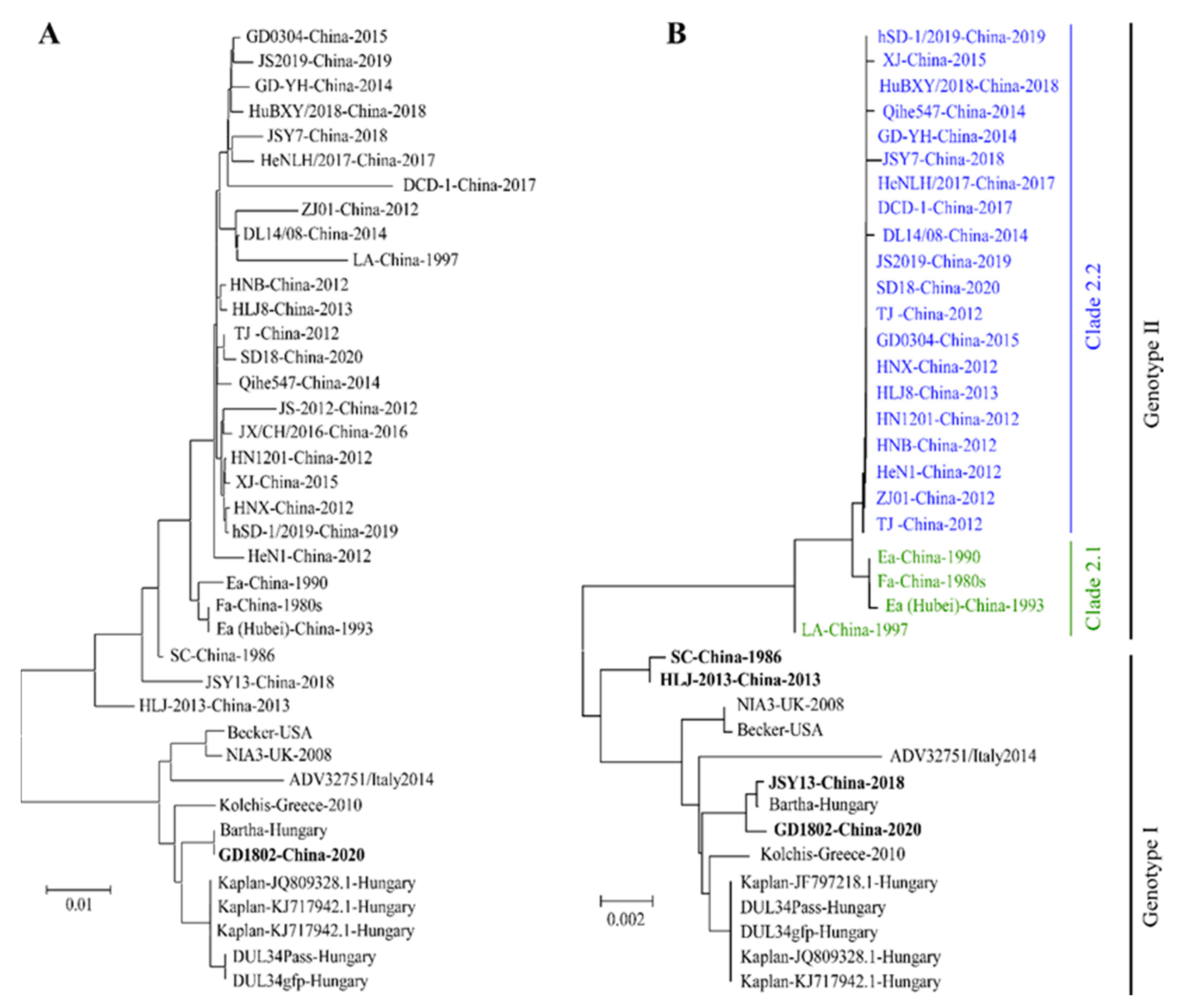
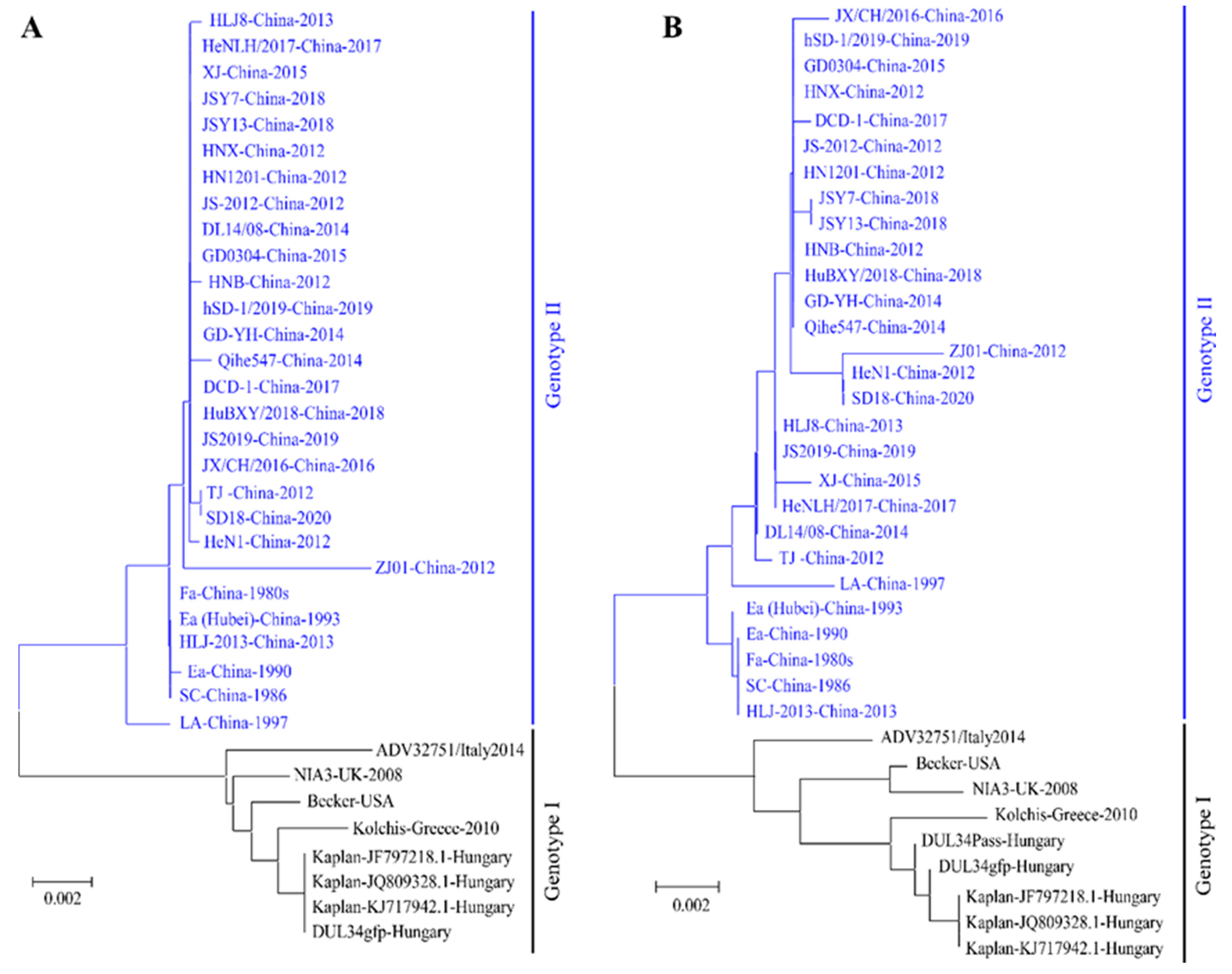
2. The Outbreaks and the Genomic Evolution of PRV Variants
3. PRV Variant-Based Vaccines Are on the Way
4. The Pathogenicity of PRV Variant to Different Species of Vertebrates
5. The Zoonotic Potential of PRV Variants
6. Discussion and Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pomeranz, L.E.; Reynolds, A.E.; Hengartner, C.J. Molecular Biology of Pseudorabies Virus: Impact on Neurovirology and Veterinary Medicine. Microbiol. Mol. Biol. Rev. 2005, 69, 462–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wittmann, G.; Rziha, H.-J. Aujeszk™ s Disease (Pseudorabies) in Pigs. In Herpesvirus Diseases of Cattle, Horses, and Pigs; Springer: Boston, MA, USA, 1989. [Google Scholar]
- Mettenleiter, T.C. Aujeszky’s disease (pseudorabies) virus: The virus and molecular pathogenesis—State of the art, June 1999. Vet. Res. 2000, 31, 99–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Q.Z.; Wu, Y.X.; Li, Y.X.; Li, Z.R.; Nan, X. The pseudorabies vaccination research. I: Pseudorabies attenuated vaccine research. Chin. J. Prev. Vet. Med. 1983, 1, 1–6. (In Chinese) [Google Scholar]
- Lomniczi, B.; Kaplan, A.S.; Ben-Porat, T. Multiple defects in the genome of pseudorabies virus can affect virulence without detectably affecting replication in cell culture. Virology 1987, 161, 181–189. [Google Scholar] [CrossRef]
- Sun, Y.; Luo, Y.; Wang, C.-H.; Yuan, J.; Li, N.; Song, K.; Qiu, H.-J. Control of swine pseudorabies in China: Opportunities and limitations. Vet. Microbiol. 2016, 183, 119–124. [Google Scholar] [CrossRef]
- Freuling, C.M.; Müller, T.F.; Mettenleiter, T.C. Vaccines against pseudorabies virus (PrV). Vet. Microbiol. 2017, 206, 3–9. [Google Scholar] [CrossRef]
- Yu, X.; Zhou, Z.; Hu, D.; Zhang, Q.; Han, T.; Li, X.; Gu, X.; Yuan, L.; Zhang, S.; Wang, B.; et al. Pathogenic Pseudorabies Virus, China, 2012. Emerg. Infect. Dis. 2014, 20, 102–104. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Auclert, L.Z.; Zhai, X.; Wong, G.; Zhang, C.; Zhu, H.; Xing, G.; Wang, S.; He, W.; Li, K.; et al. Interspecies Transmission, Genetic Diversity, and Evolutionary Dynamics of Pseudorabies Virus. J. Infect. Dis. 2019, 219, 1705–1715. [Google Scholar] [CrossRef]
- Zhai, X.; Zhao, W.; Li, K.; Zhang, C.; Wang, C.; Su, S.; Zhou, J.; Lei, J.; Xing, G.; Sun, H.; et al. Genome Characteristics and Evolution of Pseudorabies Virus Strains in Eastern China from 2017 to 2019. Virol. Sin. 2019, 34, 601–609. [Google Scholar] [CrossRef]
- Laval, K.; Enquist, L.W. The Neuropathic Itch Caused by Pseudorabies Virus. Pathogens 2020, 9, 254. [Google Scholar] [CrossRef] [Green Version]
- Müller, T.; Hahn, E.C.; Tottewitz, F.; Kramer, M.; Klupp, B.G.; Mettenleiter, T.C.; Freuling, C. Pseudorabies virus in wild swine: A global perspective. Arch. Virol. 2011, 156, 1691–1705. [Google Scholar] [CrossRef] [PubMed]
- Enquist, L.W. Life beyond eradication: Veterinary viruses in basic science. Arch. Virol. Suppl. 1999, 15, 87–109. [Google Scholar] [CrossRef] [PubMed]
- Wong, G.; Lu, J.; Zhang, W.; Gao, G.F. Pseudorabies virus: A neglected zoonotic pathogen in humans? Emerg. Microbes Infect. 2019, 8, 150–154. [Google Scholar] [CrossRef] [Green Version]
- An, T.-Q.; Peng, J.-M.; Tian, Z.-J.; Zhao, H.-Y.; Li, N.; Liu, Y.-M.; Chen, J.-Z.; Leng, C.-L.; Sun, Y.; Chang, D.; et al. Pseudorabies Virus Variant in Bartha-K61–Vaccinated Pigs, China, 2012. Emerg. Infect. Dis. 2013, 19, 1749–1755. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Tsutsui, M.; Taguchi, K.; Saitoh, A.; Muramatsu, M. Sequence variation of the gC gene among pseudorabies virus strains. Vet. Microbiol. 1996, 49, 267–272. [Google Scholar] [CrossRef]
- Ye, C.; Zhang, Q.-Z.; Tian, Z.-J.; Zheng, H.; Zhao, K.; Liu, F.; Guo, J.-C.; Tong, W.; Jiang, C.-G.; Wang, S.-J.; et al. Genomic characterization of emergent pseudorabies virus in China reveals marked sequence divergence: Evidence for the existence of two major genotypes. Virology 2015, 483, 32–43. [Google Scholar] [CrossRef]
- Zhu, Z.; Xiao, C.-T.; Fan, Y.; Cai, Z.; Lu, C.; Zhang, G.; Jiang, T.; Tan, Y.; Peng, Y. Homologous recombination shapes the genetic diversity of African swine fever viruses. Vet. Microbiol. 2019, 236, 108380. [Google Scholar] [CrossRef]
- Zhou, L.; Kang, R.; Zhang, Y.; Yu, J.; Xie, B.; Chen, C.; Li, X.; Chen, B.; Liang, L.; Zhu, J.; et al. Emergence of two novel recombinant porcine reproductive and respiratory syndrome viruses 2 (lineage 3) in Southwestern China. Vet. Microbiol. 2019, 232, 30–41. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, Y.-F. Extensive homologous recombination in classical swine fever virus: A re-evaluation of homologous recombination events in the strain AF407339. Saudi J. Biol. Sci. 2014, 21, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Wei, C.; Lin, Z.; Dai, A.; Chen, H.; Ma, Y.; Li, N.; Wu, Y.; Yang, X.; Luo, M.; Liu, J. Emergence of a novel recombinant porcine circovirus type 2 in China: PCV2c and PCV2d recombinant. Transbound. Emerg. Dis. 2019, 66, 2496–2506. [Google Scholar] [CrossRef]
- Chen, N.; Li, S.; Zhou, R.; Zhu, M.; He, S.; Ye, M.; Huang, Y.; Li, S.; Zhu, C.; Xia, P.; et al. Two novel porcine epidemic diarrhea virus (PEDV) recombinants from a natural recombinant and distinct subtypes of PEDV variants. Virus Res. 2017, 242, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Maes, R.K.; Sussman, M.D.; Vilnis, A.; Thacker, B.J. Recent developments in latency and recombination of Aujeszky’s disease (pseudorabies) virus. Vet. Microbiol. 1997, 55, 13–27. [Google Scholar] [CrossRef]
- Dangler, C.A.; Henderson, L.M.; Bowman, L.A.; Deaver, R.E. Direct isolation and identification of recombinant pseudorabies virus strains from tissues of experimentally co-infected swine. Am. J. Vet. Res. 1993, 54, 540–545. [Google Scholar] [PubMed]
- Ye, C.; Guo, J.-C.; Gao, J.-C.; Wang, T.-Y.; Zhao, K.; Chang, X.-B.; Wang, Q.; Peng, J.-M.; Tian, Z.-J.; Cai, X.-H.; et al. Genomic analyses reveal that partial sequence of an earlier pseudorabies virus in China is originated from a Bartha-vaccine-like strain. Virology 2016, 491, 56–63. [Google Scholar] [CrossRef] [PubMed]
- Bo, Z.; Miao, Y.; Xi, R.; Gao, X.; Miao, D.; Chen, H.; Jung, Y.S.; Qian, Y.; Dai, J. Emergence of a novel pathogenic recombinant virus from Bartha vaccine and variant pseudorabies virus in China. Transbound. Emerg. Dis. 2020, 68, 1454–1464. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Shi, Z.; Liu, C.; Wang, P.; Wang, M.; Wang, S.; Liu, Z.; Wei, L.; Sun, Z.; He, X.; et al. Implication of the Identification of an Earlier Pseudorabies Virus (PRV) Strain HLJ-2013 to the Evolution of Chinese PRVs. Front. Microbiol. 2020, 11, 612474. [Google Scholar] [CrossRef]
- Huang, J.; Zhu, L.; Zhao, J.; Yin, X.; Feng, Y.; Wang, X.; Sun, X.; Zhou, Y.; Xu, Z. Genetic evolution analysis of novel recombinant pseudorabies virus strain in Sichuan, China. Transbound. Emerg. Dis. 2020, 67, 1428–1432. [Google Scholar] [CrossRef]
- Ferrari, M.; Mettenleiter, T.; Romanelli, M.; Cabassi, E.; Corradi, A.; Mas, N.D.; Silini, R. A Comparative Study of Pseudorabies Virus (PRV) Strains with Defects in Thymidine Kinase and Glycoprotein Genes. J. Comp. Pathol. 2000, 123, 152–163. [Google Scholar] [CrossRef]
- Fan, J.; Zeng, X.; Zhang, G.; Wu, Q.; Niu, J.; Sun, B.; Xie, Q.; Ma, J. Molecular characterization and phylogenetic analysis of pseudorabies virus variants isolated from Guangdong province of southern China during 2013–2014. J. Vet. Sci. 2016, 17, 369–375. [Google Scholar] [CrossRef]
- Tan, L.; Yao, J.; Yang, Y.; Luo, W.; Yuan, X.; Yang, L.; Wang, A. Current Status and Challenge of Pseudorabies Virus Infection in China. Virol. Sin. 2021, 36, 588–607. [Google Scholar] [CrossRef]
- Yu, Z.-Q.; Tong, W.; Zheng, H.; Li, L.-W.; Li, G.-X.; Gao, F.; Wang, T.; Liang, C.; Ye, C.; Wu, J.-Q.; et al. Variations in glycoprotein B contribute to immunogenic difference between PRV variant JS-2012 and Bartha-K61. Vet. Microbiol. 2017, 208, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Li, N.; Cong, X.; Wang, C.-H.; Du, M.; Li, L.; Zhao, B.; Yuan, J.; Liu, D.-D.; Li, S.; et al. Pathogenicity and genomic characterization of a pseudorabies virus variant isolated from Bartha-K61-vaccinated swine population in China. Vet. Microbiol. 2014, 174, 107–115. [Google Scholar] [CrossRef] [PubMed]
- Ling, Z.; Wan-Zhu, G.; Zhi-Wen, X. Fluctuant Rule of Colostral Antibodies and the Date of Initial Immunization for the Piglet from Sows Inoculated with Pseudorabies Virus Gene-deleted Vaccine SA215. Chin. J. Vet. Sci. 2004, 24, 320–322. (In Chinese) [Google Scholar]
- He, Q.G.; Chen, H.C.; Fang, L.R.; Wu, B.; Liu, Z.F.; Xiao, S.B.; Jin, M.L. The Safety, Stablization and Immunogenicity of Double Gene-negative Mutant of Pseudorabies Virus Strain (PrV HB-98). Chin. J. Vet. Sci. 2006, 26, 165–168. (In Chinese) [Google Scholar]
- Wang, C.-H.; Yuan, J.; Qin, H.-Y.; Luo, Y.; Cong, X.; Li, Y.; Chen, J.; Li, S.; Sun, Y.; Qiu, H.-J. A novel gE-deleted pseudorabies virus (PRV) provides rapid and complete protection from lethal challenge with the PRV variant emerging in Bartha-K61-vaccinated swine population in China. Vaccine 2014, 32, 3379–3385. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, Z.; Ge, A.; Guo, R.; Qiao, Y.; Xu, M.; Wang, Z.; Liu, Y.; Zheng, Y.; Fan, H.; et al. Safety and immunogenicity of an attenuated Chinese pseudorabies variant by dual deletion of TK&gE genes. BMC Vet. Res. 2018, 14, 287. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Guo, R.; Qiao, Y.; Xu, M.; Wang, Z.; Liu, Y.; Gu, Y.; Liu, C.; Hou, J. An inactivated gE-deleted pseudorabies vaccine provides complete clinical protection and reduces virus shedding against challenge by a Chinese pseudorabies variant. BMC Vet. Res. 2016, 12, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Z.; Dong, J.; Wang, J.; Hou, C.; Sun, H.; Yang, W.; Bai, J.; Jiang, P. A novel inactivated gE/gI deleted pseudorabies virus (PRV) vaccine completely protects pigs from an emerged variant PRV challenge. Virus Res. 2015, 195, 57–63. [Google Scholar] [CrossRef]
- Tong, W.; Li, G.; Liang, C.; Liu, F.; Tian, Q.; Cao, Y.; Li, L.; Zheng, X.; Zheng, H.; Tong, G. A live, attenuated pseudorabies virus strain JS-2012 deleted for gE/gI protects against both classical and emerging strains. Antivir. Res. 2016, 130, 110–117. [Google Scholar] [CrossRef]
- Hu, R.-M.; Zhou, Q.; Song, W.-B.; Sun, E.-C.; Zhang, M.-M.; He, Q.-G.; Chen, H.-C.; Wu, B.; Liu, Z.-F. Novel pseudorabies virus variant with defects in TK, gE and gI protects growing pigs against lethal challenge. Vaccine 2015, 33, 5733–5740. [Google Scholar] [CrossRef]
- Gao, J.F.; Lai, Z.; Shu, Y.H.; Qi, S.H.; Ma, J.J.; Wu, B.Q.; Gong, J.P. Isolation and identification of porcine pseudorabies virus (PRV) C strain. Acta Agric. Shanghai 2015, 31, 32–36. (In Chinese) [Google Scholar]
- Mettenleiter, T.C. Pseudorabies (Aujeszky’s disease) virus: State of the art. August 1993. Acta Vet. Hung. 1994, 42, 153–177. [Google Scholar] [PubMed]
- Wang, Y.; Wang, T.; Yan, H.; Yang, F.; Guo, L.; Yang, Q.; Hu, X.; Tan, F.; Xiao, Y.; Li, X.; et al. Research and development of a novel subunit vaccine for the currently circulating pseudorabies virus variant in China. Front. Agric. Sci. Eng. 2015, 2, 216–222. [Google Scholar] [CrossRef]
- Zhang, T.; Liu, Y.; Chen, Y.; Wang, A.; Feng, H.; Wei, Q.; Zhou, E.; Zhang, G. A single dose glycoprotein D-based subunit vaccine against pseudorabies virus infection. Vaccine 2020, 38, 6153–6161. [Google Scholar] [CrossRef] [PubMed]
- Porter, K.R.; Raviprakash, K. DNA Vaccine Delivery and Improved Immunogenicity. Curr. Issues Mol. Biol. 2017, 22, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.M.; Haagmans, B.L.; Glansbeek, H.L.; de Visser, Y.E.; de Bruin, M.G.; Boersma, W.; Bianchi, A.T. A DNA vaccine coding for glycoprotein B of pseudorabies virus induces cell-mediated immunity in pigs and reduces virus excretion early after infection. Vet. Immunol. Immunopathol. 2000, 74, 121–136. [Google Scholar] [CrossRef]
- Yoon, H.A.; Han, Y.W.; Aleyas, A.; George, J.A.; Kim, S.J.; Kim, H.K.; Song, H.J.; Cho, J.G.; Eo, S.K. Protective immunity induced by systemic and mucosal delivery of DNA vaccine expressing glycoprotein B of pseudorabies virus. J. Microbiol. Biotechnol. 2008, 18, 591–599. [Google Scholar]
- Pardi, N.; Hogan, M.J.; Porter, F.W.; Weissman, D. mRNA vaccines—A new era in vaccinology. Nat. Rev. Drug Discov. 2018, 17, 261–279. [Google Scholar] [CrossRef] [Green Version]
- Jackson, N.A.C.; Kester, K.E.; Casimiro, D.; Gurunathan, S.; DeRosa, F. The promise of mRNA vaccines: A biotech and industrial perspective. NPJ Vaccines 2020, 5, 11. [Google Scholar] [CrossRef]
- Jiang, Z.; Zhu, L.; Cai, Y.; Yan, J.; Fan, Y.; Lv, W.; Gong, S.; Yin, X.; Yang, X.; Sun, X.; et al. Immunogenicity and protective efficacy induced by an mRNA vaccine encoding gD antigen against pseudorabies virus infection. Vet. Microbiol. 2020, 251, 108886. [Google Scholar] [CrossRef]
- Zhou, J.; Li, S.; Wang, X.; Zou, M.; Gao, S. Bartha-k61 vaccine protects growing pigs against challenge with an emerging variant pseudorabies virus. Vaccine 2017, 35, 1161–1166. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zeng, R.; Torrents, D.; Martinez, C.; Qiao, Y.; Yiqi, G.U.; Liu, C.J.A.H.; Medicine, V. Protection of pseudorabies vaccine (Bartha K61 strain) against pseudorabies virus variant in pigs. Anim. Husb. Vet. Med. 2015, 47. (In Chinese) [Google Scholar]
- Hanson, R.P. The history of pseudorabies in the United States. J. Am. Vet. Med. Assoc. 1954, 124, 259–261. [Google Scholar] [PubMed]
- Beran, G.W.; Davies, E.B.; Arambulo, P.V., 3rd; Will, L.A.; Hill, H.T.; Rock, D.L. Persistence of pseudorabies virus in infected swine. J. Am. Vet. Med. Assoc. 1980, 176, 998–1000. [Google Scholar]
- Verpoest, S.; Cay, A.B.; De Regge, N. Molecular characterization of Belgian pseudorabies virus isolates from domestic swine and wild boar. Vet. Microbiol. 2014, 172, 72–77. [Google Scholar] [CrossRef]
- Mocsári, E.; Szolnoki, J.; Glávits, R.; Zsák, L. Horizontal transmission of Aujeszky’s disease virus from sheep to pigs. Vet. Microbiol. 1989, 19, 245–252. [Google Scholar] [CrossRef]
- Egberink, H.F. Aujeszky’s disease in dogs and cats. Tijdschr. Voor Diergeneeskd. 1990, 115, 349–353. [Google Scholar]
- Raymond, J.T.; Gillespie, R.G.; Woodruff, M.; Janovitz, E.B. Pseudorabies in Captive Coyotes. J. Wildl. Dis. 1997, 33, 916–918. [Google Scholar] [CrossRef] [Green Version]
- Murdoch, R.S. Aujeszky’s disease in foxhounds. Vet. Rec. 1990, 126, 226. [Google Scholar]
- Dolivo, M.; Beretta, E.; Bonifas, V.; Foroglou, C. Ultrastructure and function in sympathetic ganglia isolated from rats infected with pseudorabies virus. Brain Res. 1978, 140, 111–123. [Google Scholar] [CrossRef]
- Neagari, Y.; Sakai, T.; Nogami, S.; Kaiho, I.; Katoh, C. Incidence of antibodies in raccoon dogs and deer inhabiting suburban areas. Kansenshogaku Zasshi 1998, 72, 331–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banks, M.; Torraca, L.S.M.; Greenwood, A.G.; Taylor, D.C. Aujeszky’s disease in captive bears. Vet. Rec. 1999, 145, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Goto, H.; Burger, D.; Gorham, J., Jr. Quantitative studies of pseudirabies virus in mink, ferrets, rabbits and mice. Jpn. J. Vet. Sci. 1971, 33, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Guillon, J.C.; Chirol, C.; Vallée, A.; Cordaillat, J.C.; Beylot, J.C. A focus of Aujeszky’s disease in dogs in the department of Ain. Bull. L’acad. Vet. Fr. 1968, 41, 177–179. [Google Scholar]
- Kimman, T.G.; Binkhorst, G.J.; Ingh, T.S.V.D.; Pol, J.M.; Gielkens, A.L.; Roelvink, M.E. Aujeszky’s disease in horses fulfils Koch’s postulates. Vet. Rec. 1991, 128, 103–106. [Google Scholar] [CrossRef]
- Reagan, R.L.; Day, W.C.; Marley, R.T.; Brueckner, A.L. Effect of pseudorabies virus (Aujeszky strain) in the large brown bat (Eptesicus fuscus). Am. J. Vet. Res. 1953, 14, 331–332. [Google Scholar]
- Amoroso, M.G.; Di Concilio, D.; D’Alessio, N.; Veneziano, V.; Galiero, G.; Fusco, G. Canine parvovirus and pseudorabies virus coinfection as a cause of death in a wolf (Canis lupus) from southern Italy. Vet. Med. Sci. 2020, 6, 600–605. [Google Scholar] [CrossRef] [Green Version]
- Thawley, D.; Wright, J. Pseudorabies virus infection in raccoons: A review. J. Wildl. Dis. 1982, 18, 113–116. [Google Scholar] [CrossRef] [Green Version]
- Field, H.J.; Hill, T.J. The Pathogenesis of Pseudorabies in Mice following Peripheral Inoculation. J. Gen. Virol. 1974, 23, 145–157. [Google Scholar] [CrossRef]
- Ohshima, K.I.; Gorham, J.R.; Henson, J.B. Pathologic changes in ferrets exposed to pseudorabies virus. Am. J. Vet. Res. 1976, 37, 591–596. [Google Scholar]
- Glass, C.M.; McLean, R.G.; Katz, J.B.; Maehr, D.S.; Cropp, C.B.; Kirk, L.J.; McKeiman, A.J.; Evermann, J.F. Isolation of pseudorabies (Aujeszky’s Disease) virus from a florida panther. J. Wildl. Dis. 1994, 30, 180–184. [Google Scholar] [CrossRef] [PubMed]
- McCracken, R.M.; McFerran, J.B.; Dow, C. The Neural Spread of Pseudorabies Virus in Calves. J. Gen. Virol. 1973, 20, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.P.; Fraser, G. Studies on the virus of Aujeszky’s disease. J. Comp. Pathol. 1971, 81, 55–62. [Google Scholar] [CrossRef]
- Tu, L.; Lian, J.; Pang, Y.; Liu, C.; Cui, S.; Lin, W. Retrospective detection and phylogenetic analysis of pseudorabies virus in dogs in China. Arch. Virol. 2021, 166, 91–100. [Google Scholar] [CrossRef]
- Cheng, Z.; Kong, Z.; Liu, P.; Fu, Z.; Zhang, J.; Liu, M.; Shang, Y. Natural infection of a variant pseudorabies virus leads to bovine death in China. Transbound. Emerg. Dis. 2020, 67, 518–522. [Google Scholar] [CrossRef]
- Lian, K.; Zhang, M.; Zhou, L.; Song, Y.; Wang, G.; Wang, S. First report of a pseudorabies-virus-infected wolf (Canis lupus) in China. Arch. Virol. 2020, 165, 459–462. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Li, X.-T.; Hu, B.; Deng, X.-Y.; Zhang, L.; Lian, S.-Z.; Zhang, H.-L.; Lv, S.; Xue, X.-H.; Lu, R.-G.; et al. Outbreak of severe pseudorabies virus infection in pig-offal-fed farmed mink in Liaoning Province, China. Arch. Virol. 2017, 162, 863–866. [Google Scholar] [CrossRef]
- Jin, H.-L.; Gao, S.-M.; Liu, Y.; Zhang, S.-F.; Hu, R.-L. Pseudorabies in farmed foxes fed pig offal in Shandong province, China. Arch. Virol. 2016, 161, 445–448. [Google Scholar] [CrossRef]
- Lin, J.; Li, Z.; Feng, Z.; Fang, Z.; Chen, J.; Chen, W.; Liang, W.; Chen, Q. Pseudorabies virus (PRV) strain with defects in gE, gC, and TK genes protects piglets against an emerging PRV variant. J. Vet. Med Sci. 2020, 82, 846–855. [Google Scholar] [CrossRef] [Green Version]
- Müller, T.F.; Teuffert, J.; Zellmer, R.; Conraths, F.J. Experimental infection of European wild boars and domestic pigs with pseudorabies viruses with differing virulence. Am. J. Vet. Res. 2001, 62, 252–258. [Google Scholar] [CrossRef]
- Schmidt, S.P.; Hagemoser, W.A.; Kluge, J.P.; Hill, H.T. Pathogenesis of ovine pseudorabies (Aujeszky’s disease) following intratracheal inoculation. Can. J. Vet. Res. 1987, 51, 326–333. [Google Scholar] [PubMed]
- Zhang, L.; Zhong, C.; Wang, J.; Lu, Z.; Liu, L.; Yang, W.; Lyu, Y. Pathogenesis of natural and experimental Pseudorabies virus infections in dogs. Virol. J. 2015, 12, 44. [Google Scholar] [CrossRef] [Green Version]
- Hagemoser, W.A.; Kluge, J.P.; Hill, H.T. Studies on the pathogenesis of pseudorabies in domestic cats following oral inoculation. Can. J. Comp. Med. 1980, 44, 192–202. [Google Scholar] [PubMed]
- Quiroga, M.I.; Vázquez, S.; López-Peña, M.; Guerrero, F.; Nieto, J.M. Experimental Aujeszky’s Disease in Blue Foxes (Alopex lagopus. J. Vet. Med. Ser. A 1995, 42, 649–657. [Google Scholar] [CrossRef] [PubMed]
- Kirkpatrick, C.M.; Kanitz, C.L.; McCrocklin, S.M. Possible Role of wild mammals in transmission of pseudorabies to swine. J. Wildl. Dis. 1980, 16, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Sehl, J.; Teifke, J.P. Comparative Pathology of Pseudorabies in Different Naturally and Experimentally Infected Species—A Review. Pathogens 2020, 9, 633. [Google Scholar] [CrossRef]
- Laval, K.; Vernejoul, J.B.; Van Cleemput, J.; Koyuncu, O.O.; Enquist, L.W. Virulent Pseudorabies Virus Infection Induces a Specific and Lethal Systemic Inflammatory Response in Mice. J. Virol. 2018, 92, e01614–e01618. [Google Scholar] [CrossRef] [Green Version]
- Rassnick, S.; Enquist, L.W.; Sved, A.F.; Card, J. Pseudorabies Virus-Induced Leukocyte Trafficking into the Rat Central Nervous System. J. Virol. 1998, 72, 9181–9191. [Google Scholar] [CrossRef]
- Olander, H.J.; Saunders, J.; Gustafson, D.; Jones, R. Pathologic Findings in Swine Affected with a Virulent Strain of Aujeszky’s Virus. Pathol. Vet. 1966, 3, 64–82. [Google Scholar] [CrossRef]
- Shope, R.E. Modification of the pathogenicity of pseudorabies virus by animal passage. J. Exp. Med. 1933, 57, 925–931. [Google Scholar] [CrossRef] [Green Version]
- Hurst, E.W. Studies on pseudorabies (infectious bulbar paralysis, mad itch). J. Exp. Med. 1936, 63, 449–463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Liang, R.; Pang, Y.; Shi, L.; Cui, S.; Lin, W. Evidence for interspecies transmission route of pseudorabies virus via virally contaminated fomites. Vet. Microbiol. 2020, 251, 108912. [Google Scholar] [CrossRef] [PubMed]
- Hahn, E.; Page, G.; Hahn, P.; Gillis, K.; Romero, C.; Annelli, J.; Gibbs, E. Mechanisms of transmission of Aujeszky’s disease virus originating from feral swine in the USA. Vet. Microbiol. 1997, 55, 123–130. [Google Scholar] [CrossRef]
- Reilly, L.M.; Rall, G.; Lomniczi, B.; Mettenleiter, T.C.; Kuperschmidt, S.; Ben-Porat, T. The ability of pseudorabies virus to grow in different hosts is affected by the duplication and translocation of sequences from the left end of the genome to the UL-US junction. J. Virol. 1991, 65, 5839–5847. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.; Yan, J.; Lu, G.; Guo, Z.; Fan, Z.; Wang, J.; Shi, Y.; Qi, J.; Gao, G.F. Binding of herpes simplex virus glycoprotein D to nectin-1 exploits host cell adhesion. Nat. Commun. 2011, 2, 577. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.-W.; Weng, S.-S.; Cheng, Q.; Cui, P.; Li, Y.-J.; Wu, H.-L.; Zhu, Y.-M.; Xu, B.; Zhang, W.-H. Human Endophthalmitis Caused By Pseudorabies Virus Infection, China, 2017. Emerg. Infect. Dis. 2018, 24, 1087–1090. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Wang, X.; Xie, C.; Ding, S.; Yang, H.; Guo, S.; Li, J.; Qin, L.; Ban, F.; Wang, D.; et al. A Novel Human Acute Encephalitis Caused by Pseudorabies Virus Variant Strain. Clin. Infect. Dis. 2021, 73, e3690–e3700. [Google Scholar] [CrossRef]
- Skinner, G.; Ahmad, A.; Davies, J. The infrequency of transmission of herpesviruses between humans and animals; postulation of an unrecognised protective host mechanism. Comp. Immunol. Microbiol. Infect. Dis. 2001, 24, 255–269. [Google Scholar] [CrossRef]
- Avak, S.; Bienzle, U.; Feldmeier, H.; Hampl, H.; Habermehl, K.-O. Pseudorabies in man. Lancet 1987, 1, 501–502. [Google Scholar] [CrossRef]
- Anusz, Z.; Szweda, W.; Popko, J.; Trybała, E. Is Aujeszky’s disease a zoonosis? Prz. Epidemiol. 1992, 46, 181–186. [Google Scholar]
- Zhao, W.L.; Wu, Y.H.; Li, H.F.; Li, S.Y.; Fan, S.Y.; Wu, H.L.; Li, Y.J.; Lü, Y.L.; Han, J.; Zhang, W.C.; et al. Clinical experience and next-generation sequencing analysis of encephalitis caused by pseudorabies virus. Zhonghua Yi Xue Za Zhi 2018, 98, 1152–1157. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Guan, H.; Li, C.; Li, Y.; Wang, S.; Zhao, X.; Zhao, Y.; Liu, Y. Characteristics of human encephalitis caused by pseudorabies virus: A case series study. Int. J. Infect. Dis. 2019, 87, 92–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Nian, H.; Li, Z.; Wang, W.; Wang, X.; Cui, Y. Human encephalitis complicated with bilateral acute retinal necrosis associated with pseudorabies virus infection: A case report. Int. J. Infect. Dis. 2019, 89, 51–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Han, H.; Wang, H.; Cui, Y.; Liu, H.; Ding, S. A Case of Human Viral Encephalitis Caused by Pseudorabies Virus Infection in China. Front. Neurol. 2019, 10, 534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Liu, X.; Yuan, D.; Li, R.; Lu, J.; Li, X.; Tian, K.; Dai, E. Dynamic cerebrospinal fluid analyses of severe pseudorabies encephalitis. Transbound. Emerg. Dis. 2019, 66, 2562–2565. [Google Scholar] [CrossRef]
- Wang, D.; Tao, X.; Fei, M.; Chen, J.; Guo, W.; Li, P.; Wang, J. Human encephalitis caused by pseudorabies virus infection: A case report. J. Neuro Virol. 2020, 26, 442–448. [Google Scholar] [CrossRef]
- Fan, S.; Yuan, H.; Liu, L.; Li, H.; Wang, S.; Zhao, W.; Wu, Y.; Wang, P.; Hu, Y.; Han, J.; et al. Pseudorabies virus encephalitis in humans: A case series study. J. Neuro Virol. 2020, 26, 556–564. [Google Scholar] [CrossRef]
- Hu, F.; Wang, J.; Peng, X.-Y. Bilateral Necrotizing Retinitis following Encephalitis Caused by the Pseudorabies Virus Confirmed by Next-Generation Sequencing. Ocul. Immunol. Inflamm. 2021, 29, 922–925. [Google Scholar] [CrossRef]
- Ying, M.; Hu, X.; Wang, M.; Cheng, X.; Zhao, B.; Tao, Y. Vitritis and retinal vasculitis caused by pseudorabies virus. J. Int. Med. Res. 2021, 49, 3000605211058990. [Google Scholar] [CrossRef]
- Yan, W.; Hu, Z.; Zhang, Y.; Wu, X.; Zhang, H. Case Report: Metagenomic Next-Generation Sequencing for Diagnosis of Human Encephalitis and Endophthalmitis Caused by Pseudorabies Virus. Front. Med. 2021, 8, 753988. [Google Scholar] [CrossRef]
- Zhou, Y.; Nie, C.; Wen, H.; Long, Y.; Zhou, M.; Xie, Z.; Hong, D. Human viral encephalitis associated with suid herpesvirus 1. Neurol. Sci. 2022, 43, 2681–2692. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Cui, X.; Wang, X.; Wang, W.; Gao, S.; Liu, X.; Kai, Y.; Chen, C. Efficacy of the Bartha-K61 vaccine and a gE−/gI−/TK− prototype vaccine against variant porcine pseudorabies virus (vPRV) in piglets with sublethal challenge of vPRV. Res. Vet. Sci. 2020, 128, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Ketusing, N.; Reeves, A.; Portacci, K.; Yano, T.; Olea-Popelka, F.; Keefe, T.; Salman, M. Evaluation of Strategies for the Eradication of Pseudorabies Virus (Aujeszky’s Disease) in Commercial Swine Farms in Chiang-Mai and Lampoon Provinces, Thailand, Using a Simulation Disease Spread Model. Transbound. Emerg. Dis. 2014, 61, 169–176. [Google Scholar] [CrossRef] [PubMed]
- Ren, Q.; Ren, H.; Gu, J.; Wang, J.; Jiang, L.; Gao, S. The Epidemiological Analysis of Pseudorabies Virus and Pathogenicity of the Variant Strain in Shandong Province. Front. Vet. Sci. 2022, 9, 806824. [Google Scholar] [CrossRef]
- Zheng, H.-H.; Jin, Y.; Hou, C.-Y.; Li, X.-S.; Zhao, L.; Wang, Z.-Y.; Chen, H.-Y. Seroprevalence investigation and genetic analysis of pseudorabies virus within pig populations in Henan province of China during 2018–2019. Infect. Genet. Evol. 2021, 92, 104835. [Google Scholar] [CrossRef]
- Lin, Y.; Tan, L.; Wang, C.; He, S.; Fang, L.; Wang, Z.; Zhong, Y.; Zhang, K.; Liu, D.; Yang, Q.; et al. Serological Investigation and Genetic Characteristics of Pseudorabies Virus in Hunan Province of China From 2016 to 2020. Front. Vet. Sci. 2021, 8, 762326. [Google Scholar] [CrossRef]
- He, W.; Zhai, X.; Su, J.; Ye, R.; Zheng, Y.; Su, S. Antiviral Activity of Germacrone against Pseudorabies Virus in Vitro. Pathogens 2019, 8, 258. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.; Hu, D.; Yuan, L.; Lian, Z.; Yao, X.; Zhu, Z.; Nowotny, N.; Shi, Y.; Li, X. Meclizine Inhibits Pseudorabies Virus Replication by Interfering with Virus Entry and Release. Front. Microbiol. 2021, 12, 795593. [Google Scholar] [CrossRef]
- Qiu, H.-J.; Tian, Z.-J.; Tong, G.-Z.; Zhou, Y.-J.; Ni, J.-Q.; Luo, Y.-Z.; Cai, X.-H. Protective immunity induced by a recombinant pseudorabies virus expressing the GP5 of porcine reproductive and respiratory syndrome virus in piglets. Vet. Immunol. Immunopathol. 2005, 106, 309–319. [Google Scholar] [CrossRef]
- Tian, Z.-J.; Zhou, G.-H.; Zheng, B.-L.; Qiu, H.-J.; Ni, J.-Q.; Yang, H.-L.; Yin, X.-N.; Hu, S.-P.; Tong, G.-Z. A recombinant pseudorabies virus encoding the HA gene from H3N2 subtype swine influenza virus protects mice from virulent challenge. Vet. Immunol. Immunopathol. 2006, 111, 211–218. [Google Scholar] [CrossRef]
- Zhang, C.; Guo, S.; Guo, R.; Chen, S.; Zheng, Y.; Xu, M.; Wang, Z.; Liu, Y.; Wang, J. Identification of four insertion sites for foreign genes in a pseudorabies virus vector. BMC Vet. Res. 2021, 17, 190. [Google Scholar] [CrossRef] [PubMed]

| Year | Numbers | Symptoms | Contacted Animals | Ref. |
|---|---|---|---|---|
| 1914 | 2 | Swelling, reddening, intense itching | Cat | [99] |
| 1940 | 2 | Pruritis, erythema, and pain around the wound | Dog | [99] |
| 1963 | 2 | Throat pain, weakness in the legs, | Dog | [99] |
| 1983 | 1 | Pain in togue, hypersalivation, dysphagia, headache, arthralgia | Cat | [100] |
| 1986 | 2 | Tension in the mouth, nose, and throat; perception of strange smells and taste | Cat, or other domestic animals | [100] |
| 1992 | 6 | Pruritus in the palms, lower and upper arms, shoulders, and back | Cow | [101] |
| 2017 | 1 | Endophthalmitis, fever, headaches | Pig | [97] |
| 2018 | 4 | Encephalitis | Pig | [102] |
| 2019 | 5 | Encephalitis | Pig | [103] |
| 2019 | 1 | Encephalitis | Pig | [104] |
| 2019 | 1 | Encephalitis, fever, headache, seizure | unknown | [105] |
| 2019 | 1 | Encephalitis | Pig | [106] |
| 2020 | 1 | Encephalitis | Pig | [107] |
| 2020 | 6 | panencephalitis | Pig | [108] |
| 2021 | 1 | Retinitis | Pig | [109] |
| 2021 | 1 | Encephalitis, retinal vasculitis, fever | Pig | [110] |
| 2021 | 1 | Encephalitis, Endophthalmitis | Pig | [111] |
| 2022 | 2 | Encephalitis, seizures, endophthalmitis | Pig | [112] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bo, Z.; Li, X. A Review of Pseudorabies Virus Variants: Genomics, Vaccination, Transmission, and Zoonotic Potential. Viruses 2022, 14, 1003. https://doi.org/10.3390/v14051003
Bo Z, Li X. A Review of Pseudorabies Virus Variants: Genomics, Vaccination, Transmission, and Zoonotic Potential. Viruses. 2022; 14(5):1003. https://doi.org/10.3390/v14051003
Chicago/Turabian StyleBo, Zongyi, and Xiangdong Li. 2022. "A Review of Pseudorabies Virus Variants: Genomics, Vaccination, Transmission, and Zoonotic Potential" Viruses 14, no. 5: 1003. https://doi.org/10.3390/v14051003






