Expression of NAC1 Restrains the Memory Formation of CD8+ T Cells during Viral Infection
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Experiments
2.2. T Cell Isolation, T Cell Activation, and T Cell Culture
2.3. Virus Preparation and Titration
2.4. Viral Infection
2.5. Western Blot
2.6. Memory T Cell Tetramer Staining and Flow Cytometry
2.7. RNA Extraction, cDNA Synthesis, and qPCR
2.8. Seahorse Assay
2.9. CHIP-Seq
2.10. CFSE Labeling
2.11. Statistical Analysis
3. Results
3.1. Generation of VACV for Mouse Infection
3.2. Defects in the Survival of NAC1−/− CD8+ T Cells
3.3. Defects in Glycolysis and Oxidative Phosphorylation Rate of NAC1−/− CD8+ T Cells
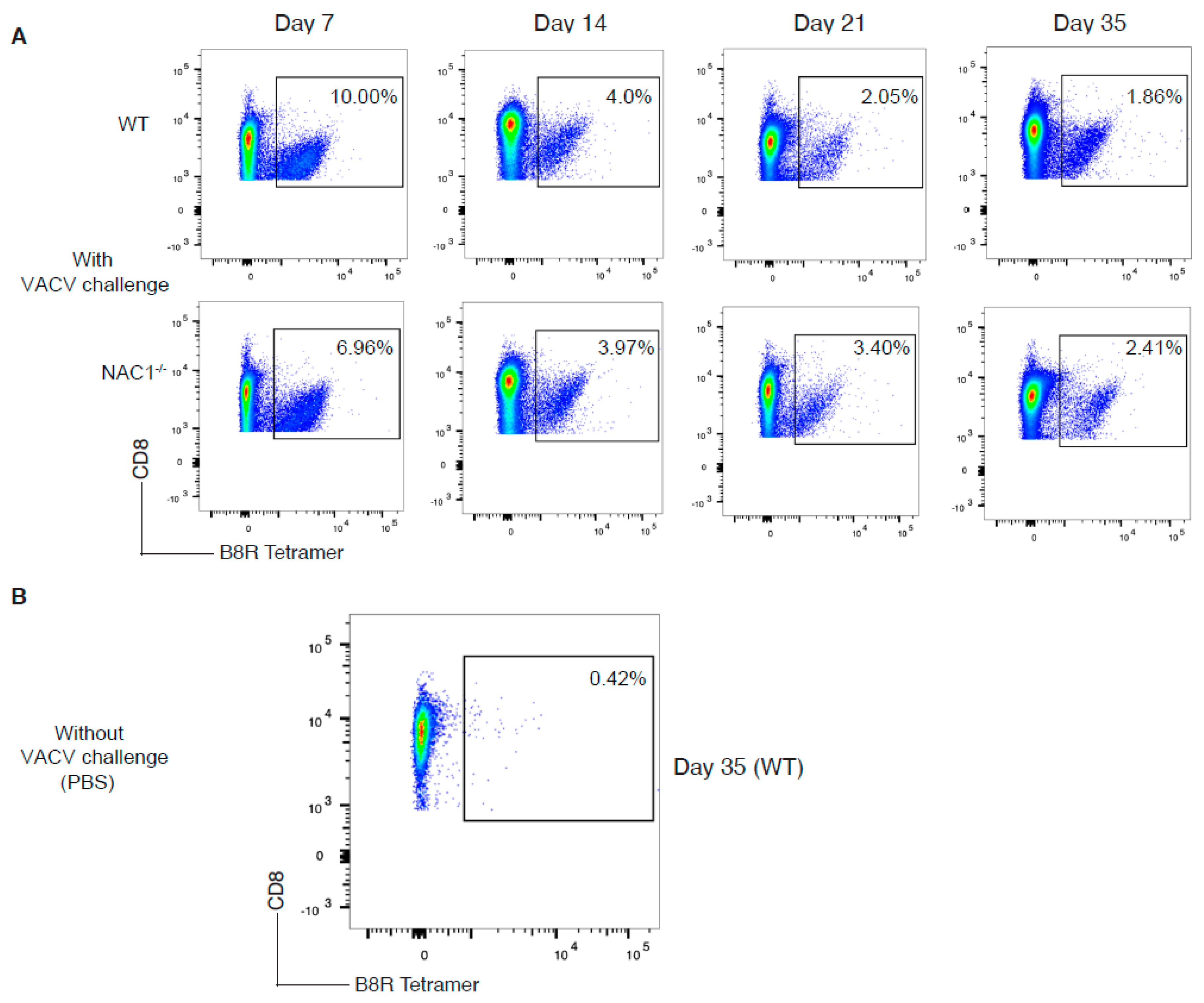
3.4. Sustained Survival of the VACV-Specific CD8+ T Cell in NAC1−/− Mice
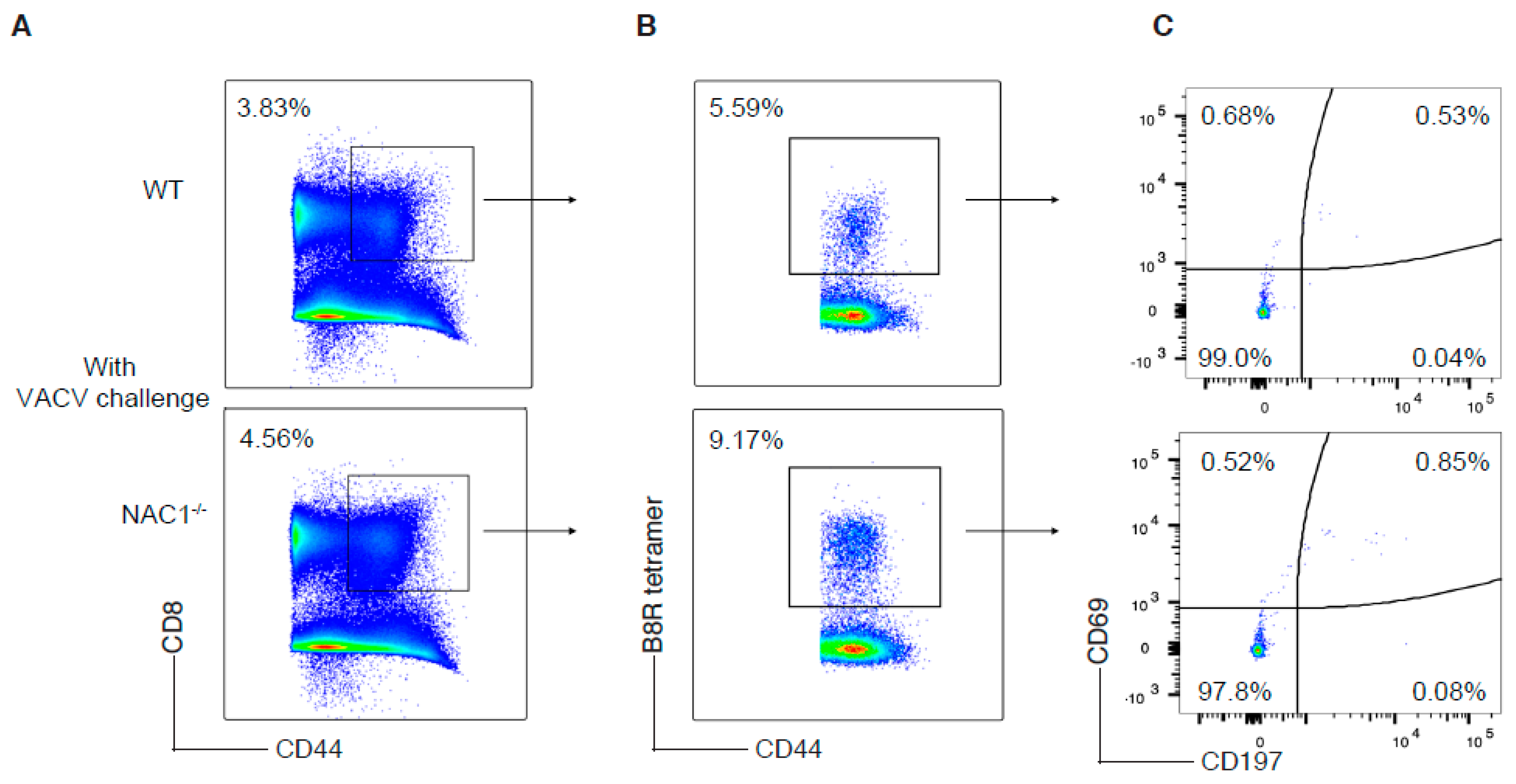
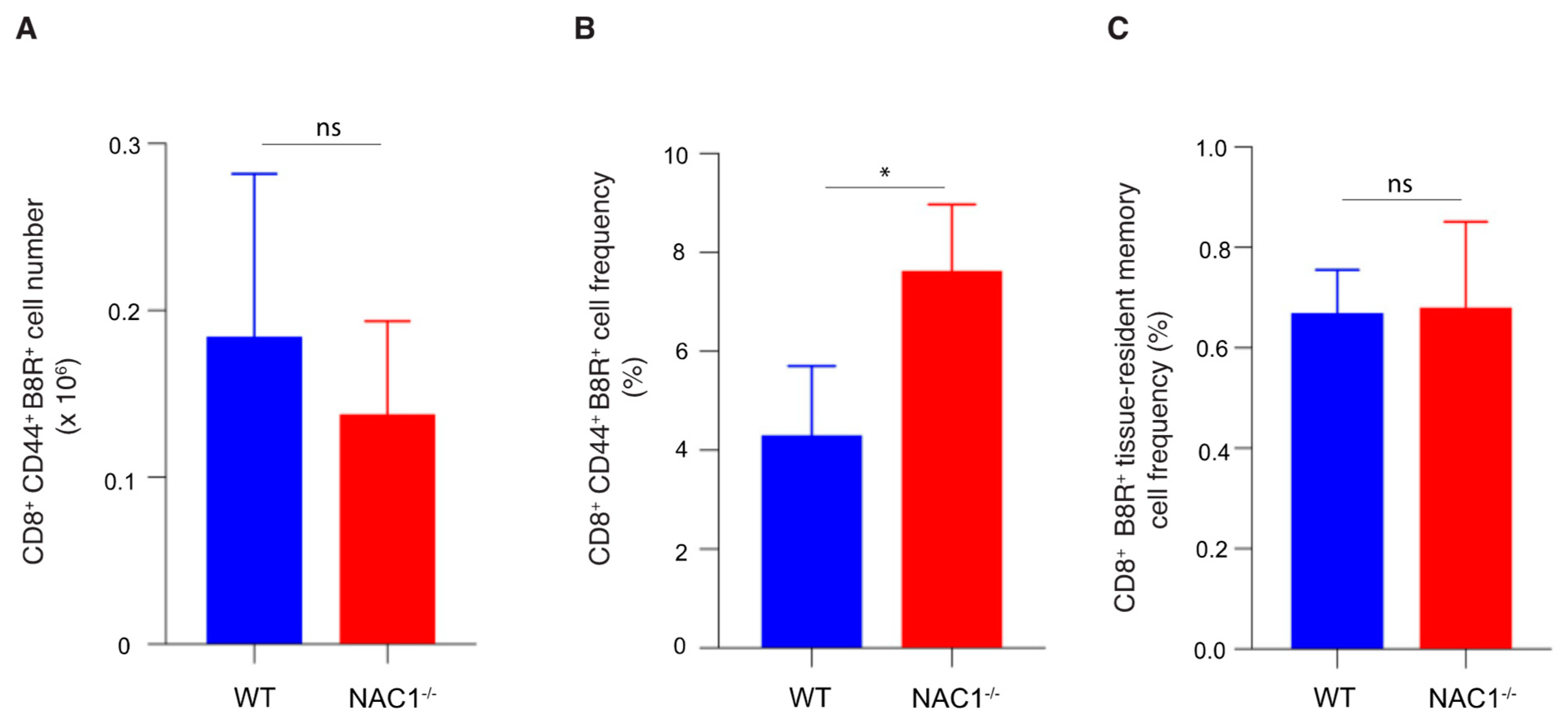
3.5. Enhanced CD8+ T Cell Memory Formation in NAC1−/− Mice
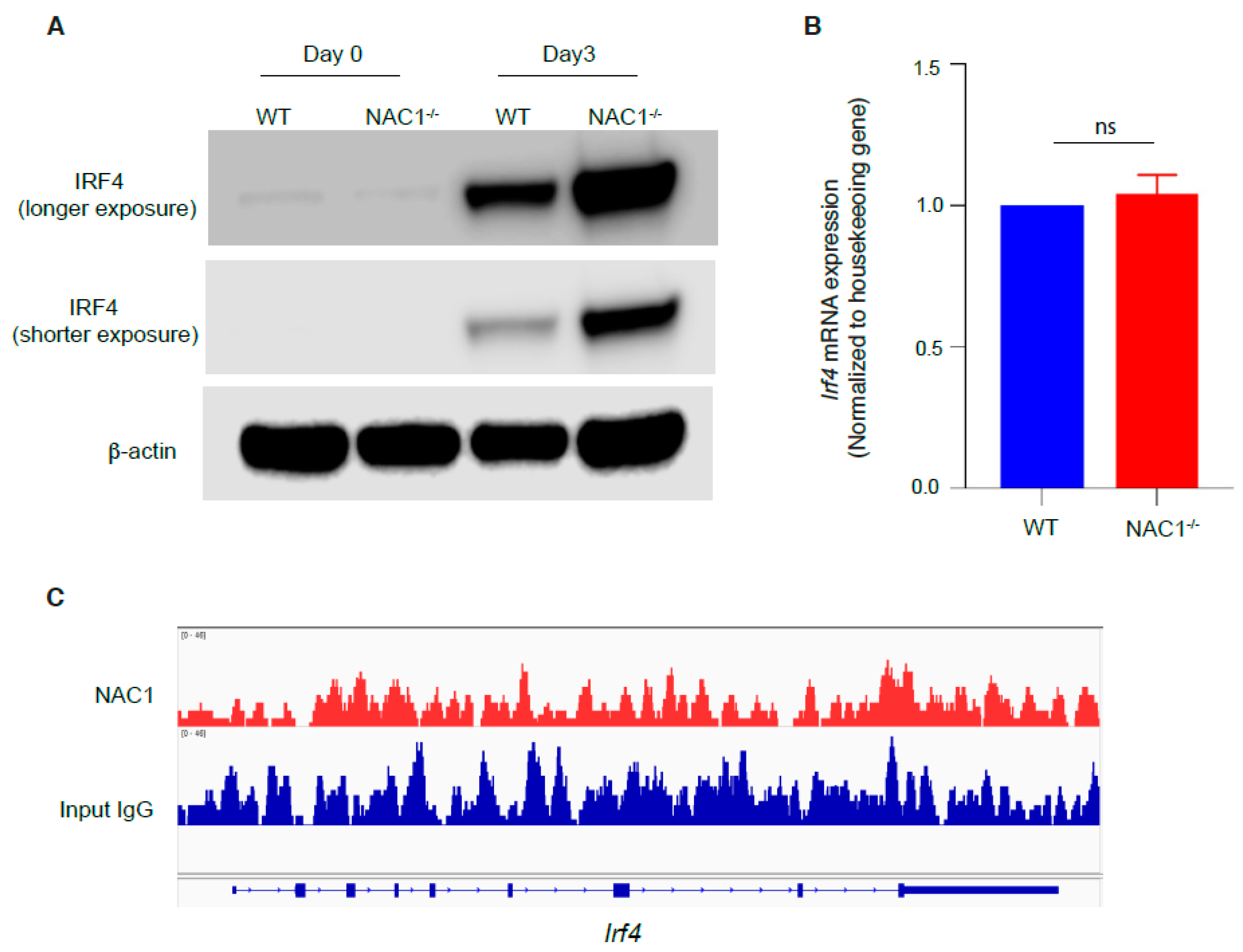
3.6. Involvement of IRF4 in the NAC1-Mediated Restrain of CD8+ T Memory Formation
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pennock, N.D.; White, J.T.; Cross, E.W.; Cheney, E.E.; Tamburini, B.A.; Kedl, R.M. T cell responses: Naive to memory and everything in between. Adv. Physiol. Educ. 2013, 37, 273–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Selin, L.K.; Welsh, R.M. Plasticity of T cell memory responses to viruses. Immunity 2004, 20, 5–16. [Google Scholar] [CrossRef] [Green Version]
- Shepherd, F.R.; McLaren, J.E. T Cell Immunity to Bacterial Pathogens: Mechanisms of Immune Control and Bacterial Evasion. Int. J. Mol. Sci. 2020, 21, 6144. [Google Scholar] [CrossRef]
- Wherry, E.J.; Ahmed, R. Memory CD8 T-cell differentiation during viral infection. J. Virol. 2004, 78, 5535–5545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klebanoff, C.A.; Gattinoni, L.; Restifo, N.P. CD8+ T-cell memory in tumor immunology and immunotherapy. Immunol. Rev. 2006, 211, 214–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Redmond, W.L.; Ruby, C.E.; Weinberg, A.D. The role of OX40-mediated co-stimulation in T-cell activation and survival. Crit. Rev. Immunol. 2009, 29, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Haque, M.; Song, J.; Fino, K.; Wang, Y.; Sandhu, P.; Song, X.; Norbury, C.; Ni, B.; Fang, D.; Salek-Ardakani, S.; et al. C-Myc regulation by costimulatory signals modulates the generation of CD8+ memory T cells during viral infection. Open Biol. 2016, 6, 150208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tejera, M.M.; Kim, E.H.; Sullivan, J.A.; Plisch, E.H.; Suresh, M. FoxO1 controls effector-to-memory transition and maintenance of functional CD8 T cell memory. J. Immunol. 2013, 191, 187–199. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Shan, Q.; Xue, H.H. TCF1 in T cell immunity: A broadened frontier. Nat. Rev. Immunol. 2021, 22, 147–157. [Google Scholar] [CrossRef]
- Korutla, L.; Wang, P.; Jackson, T.G.; Mackler, S.A. NAC1, a POZ/BTB protein that functions as a corepressor. Neurochem. Int. 2009, 54, 245–252. [Google Scholar] [CrossRef]
- Malleshaiah, M.; Padi, M.; Rue, P.; Quackenbush, J.; Martinez-Arias, A.; Gunawardena, J. Nac1 Coordinates a Sub-network of Pluripotency Factors to Regulate Embryonic Stem Cell Differentiation. Cell Rep. 2016, 14, 1181–1194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Cheng, Y.; Ren, X.; Zhang, L.; Yap, K.L.; Wu, H.; Patel, R.; Liu, D.; Qin, Z.H.; Shih, I.M.; et al. NAC1 modulates sensitivity of ovarian cancer cells to cisplatin by altering the HMGB1-mediated autophagic response. Oncogene 2012, 31, 1055–1064. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Cheng, Y.; Ren, X.; Hori, T.; Huber-Keener, K.J.; Zhang, L.; Yap, K.L.; Liu, D.; Shantz, L.; Qin, Z.H.; et al. Dysfunction of nucleus accumbens-1 activates cellular senescence and inhibits tumor cell proliferation and oncogenesis. Cancer Res. 2012, 72, 4262–4275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinawath, N.; Vasoontara, C.; Yap, K.L.; Thiaville, M.M.; Nakayama, K.; Wang, T.L.; Shih, I.M. NAC-1, a potential stem cell pluripotency factor, contributes to paclitaxel resistance in ovarian cancer through inactivating Gadd45 pathway. Oncogene 2009, 28, 1941–1948. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Ren, Y.J.; Guo, L.C.; Ji, C.; Hu, J.; Zhang, H.H.; Xu, Q.H.; Zhu, W.D.; Ming, Z.J.; Yuan, Y.S.; et al. Nucleus accumbens-associated protein-1 promotes glycolysis and survival of hypoxic tumor cells via the HDAC4-HIF-1α axis. Oncogene 2017, 36, 4171–4181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.M.; Ren, Y.; Kumar, A.; Xiong, X.; Das, J.K.; Peng, H.Y.; Wang, L.; Ren, X.; Zhang, Y.; Ji, C.; et al. NAC1 modulates autoimmunity by suppressing regulatory T cell-mediated tolerance. Sci. Adv. 2022, 8, eabo0183. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.; Lohoff, M. IRF4 at the crossroads of effector T-cell fate decision. Eur. J. Immunol. 2014, 44, 1886–1895. [Google Scholar] [CrossRef]
- Harberts, A.; Schmidt, C.; Schmid, J.; Reimers, D.; Koch-Nolte, F.; Mittrücker, H.W.; Raczkowski, F. Interferon regulatory factor 4 controls effector functions of CD8(+) memory T cells. Proc. Natl. Acad. Sci. USA 2021, 118, e2014553118. [Google Scholar] [CrossRef]
- Cotter, C.A.; Earl, P.L.; Wyatt, L.S.; Moss, B. Preparation of Cell Cultures and Vaccinia Virus Stocks. Curr. Protoc. Mol. Biol. 2017, 117, 16.16.1–16.16.18. [Google Scholar] [CrossRef] [Green Version]
- Salek-Ardakani, S.; Moutaftsi, M.; Crotty, S.; Sette, A.; Croft, M. OX40 drives protective vaccinia virus-specific CD8 T cells. J. Immunol. 2008, 181, 7969–7976. [Google Scholar] [CrossRef]
- Wong, Y.C.; Croft, S.; Smith, S.A.; Lin, L.C.W.; Cukalac, T.; La Gruta, N.L.; Drexler, I.; Tscharke, D.C. Modified Vaccinia Virus Ankara Can Induce Optimal CD8(+) T Cell Responses to Directly Primed Antigens Depending on Vaccine Design. J. Virol. 2019, 93, e01154-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Ji, C.; Zhang, H.; Shan, Y.; Ren, Y.; Hu, Y.; Shi, L.; Guo, L.; Zhu, W.; Xia, Y.; et al. Identification of a small-molecule compound that inhibits homodimerization of oncogenic NAC1 protein and sensitizes cancer cells to anticancer agents. J. Biol. Chem. 2019, 294, 10006–10017. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Jin, X.; Sun, R.; Xiong, X.; Wang, J.; Xie, D.; Zhao, M. Optimization of metabolism to improve efficacy during CAR-T cell manufacturing. J. Transl. Med. 2021, 19, 499. [Google Scholar] [CrossRef] [PubMed]
- Tartof, S.Y.; Slezak, J.M.; Fischer, H.; Hong, V.; Ackerson, B.K.; Ranasinghe, O.N.; Frankland, T.B.; Ogun, O.A.; Zamparo, J.M.; Gray, S.; et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: A retrospective cohort study. Lancet 2021, 398, 1407–1416. [Google Scholar] [CrossRef]
- Thomas, S.J.; Moreira, E.D., Jr.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Polack, F.P.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine through 6 Months. N. Engl. J. Med. 2021, 385, 1761–1773. [Google Scholar] [CrossRef] [PubMed]
- Man, K.; Gabriel, S.S.; Liao, Y.; Gloury, R.; Preston, S.; Henstridge, D.C.; Pellegrini, M.; Zehn, D.; Berberich-Siebelt, F.; Febbraio, M.A.; et al. Transcription Factor IRF4 Promotes CD8(+) T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity 2017, 47, 1129–1141.e5. [Google Scholar] [CrossRef] [Green Version]
- Raczkowski, F.; Ritter, J.; Heesch, K.; Schumacher, V.; Guralnik, A.; Höcker, L.; Raifer, H.; Klein, M.; Bopp, T.; Harb, H.; et al. The transcription factor Interferon Regulatory Factor 4 is required for the generation of protective effector CD8+ T cells. Proc. Natl. Acad. Sci. USA 2013, 110, 15019–15024. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Kumar, A.; Das, J.K.; Ren, Y.; Peng, H.-Y.; Ballard, D.J.; Xiong, X.; Davis, J.R.; Ren, X.; Yang, J.-M.; et al. Expression of NAC1 Restrains the Memory Formation of CD8+ T Cells during Viral Infection. Viruses 2022, 14, 1713. https://doi.org/10.3390/v14081713
Wang L, Kumar A, Das JK, Ren Y, Peng H-Y, Ballard DJ, Xiong X, Davis JR, Ren X, Yang J-M, et al. Expression of NAC1 Restrains the Memory Formation of CD8+ T Cells during Viral Infection. Viruses. 2022; 14(8):1713. https://doi.org/10.3390/v14081713
Chicago/Turabian StyleWang, Liqing, Anil Kumar, Jugal Kishore Das, Yijie Ren, Hao-Yun Peng, Darby Jane Ballard, Xiaofang Xiong, Jacob Rance Davis, Xingcong Ren, Jin-Ming Yang, and et al. 2022. "Expression of NAC1 Restrains the Memory Formation of CD8+ T Cells during Viral Infection" Viruses 14, no. 8: 1713. https://doi.org/10.3390/v14081713






