Virus Diversity, Abundance, and Evolution in Three Different Bat Colonies in Switzerland
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Type
2.2. Sample Preparation and Collection
2.3. Virome Analysis
2.3.1. Comparison of Viruses and Genes
2.3.2. Test for Shifts in SNV Frequency
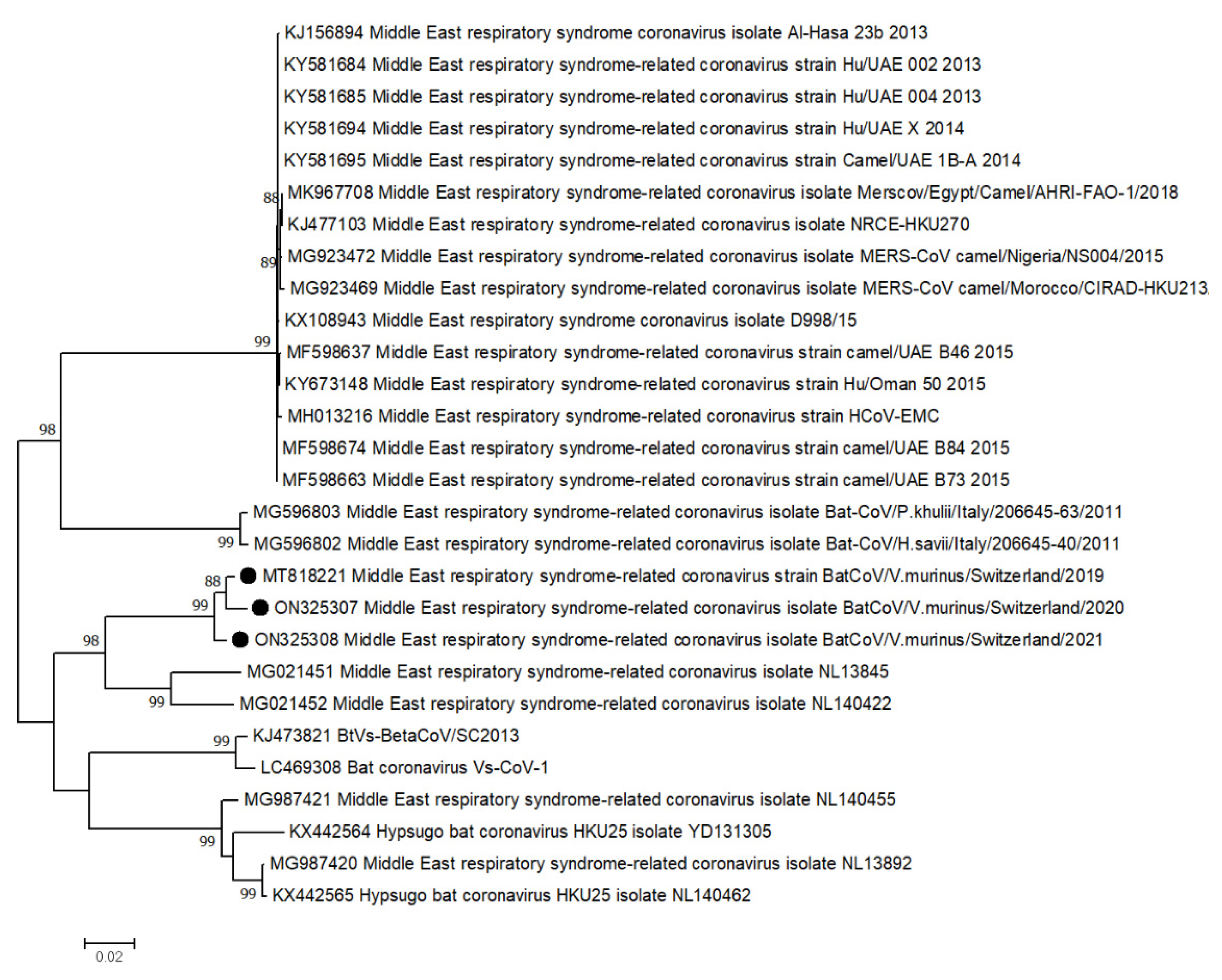
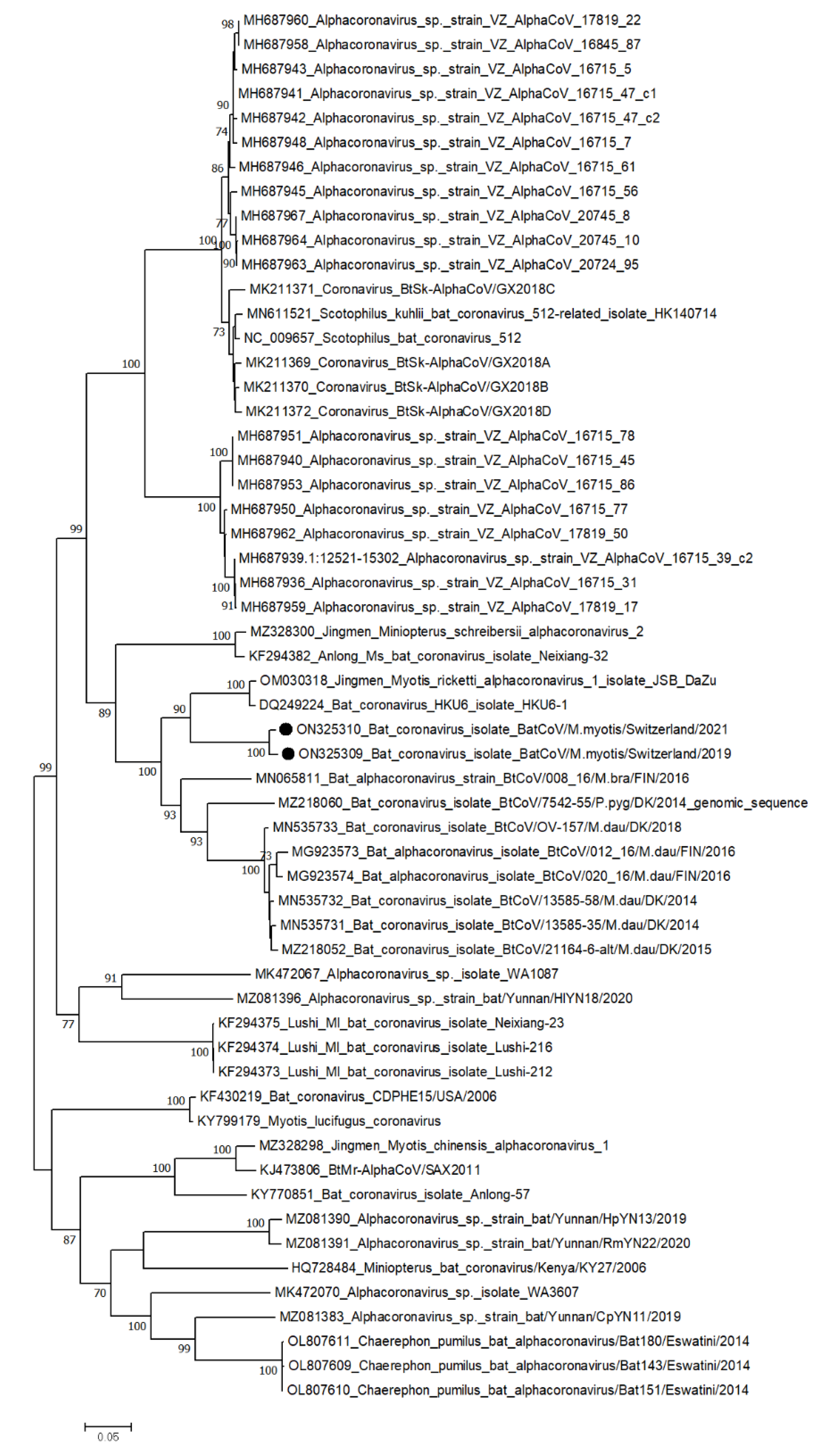
2.4. Phylogenetic Analysis
2.5. PCR
2.6. Data Availability
3. Results
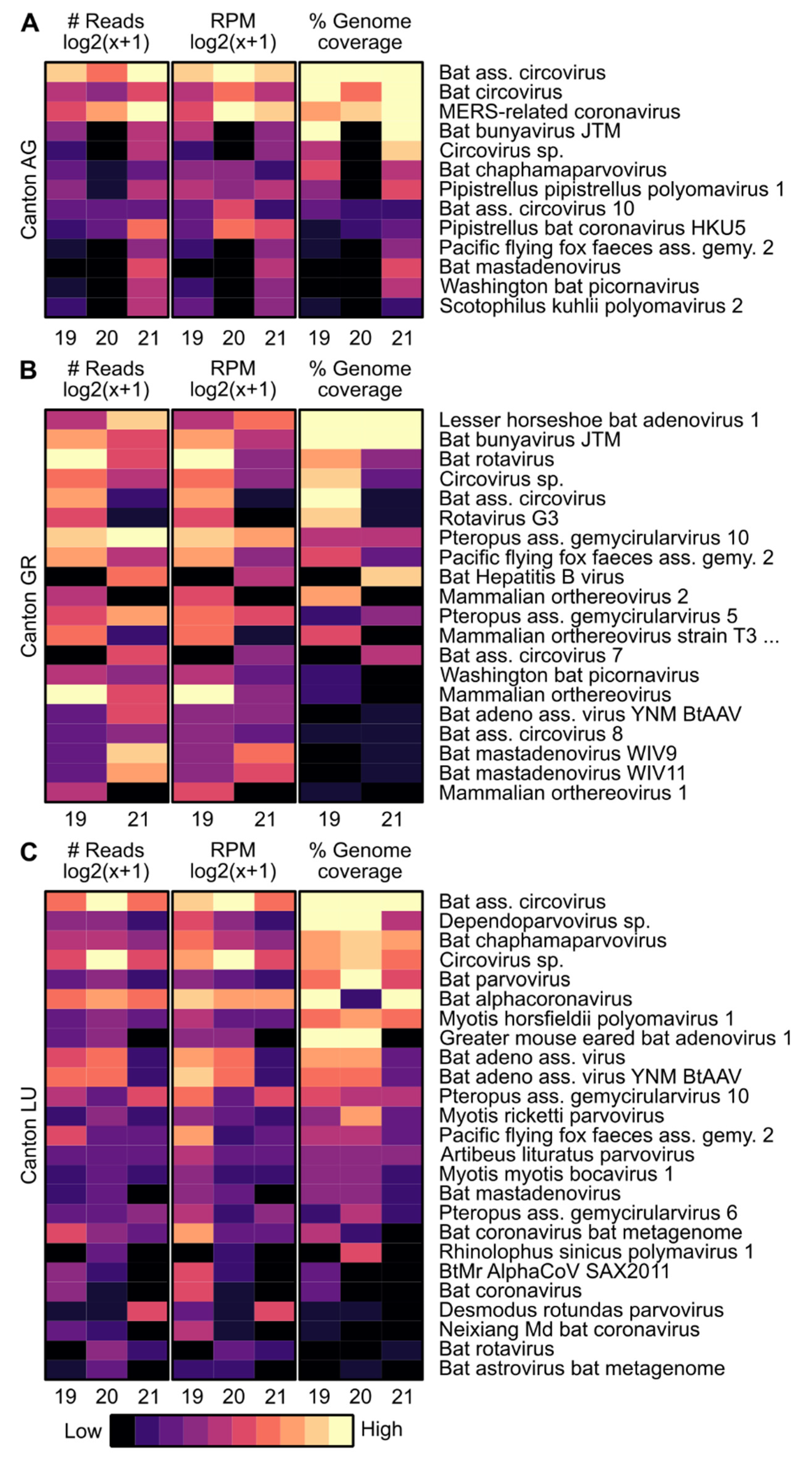
3.1. Virome Analysis
3.2. Variability of Selected Genomes and ORFs over Time
3.2.1. Variability of Selected DNA Virus Genomes
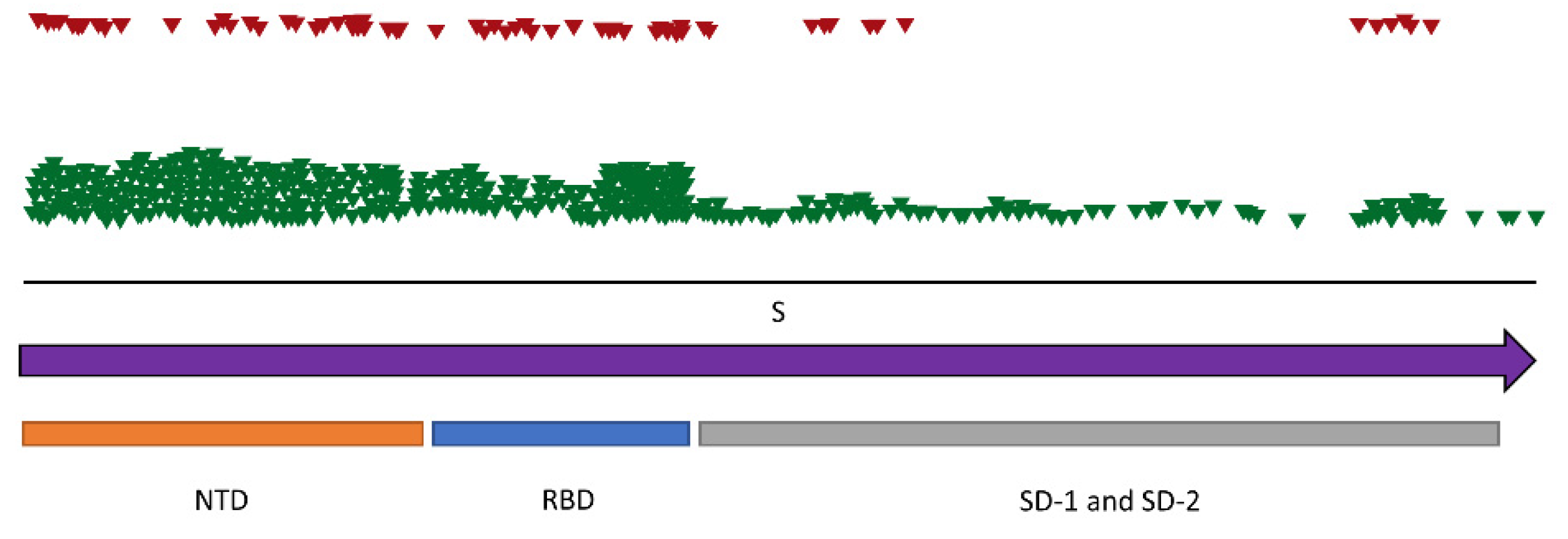
3.2.2. Variability of Selected RNA Virus Genomes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jones, K.E.; Patel, N.G.; Levy, M.A.; Storeygard, A.; Balk, D.; Gittleman, J.L.; Daszak, P. Global trends in emerging infectious diseases. Nature 2008, 451, 990–993. [Google Scholar] [CrossRef] [PubMed]
- Smith, I.; Wang, L.F. Bats and their virome: An important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 2013, 3, 84–91. [Google Scholar] [CrossRef] [PubMed]
- Williams, E.S.; Yuill, T.; Artois, M.; Fischer, J.; Haigh, S.A. Emerging infectious diseases in wildlife. Rev. Sci. Tech. 2002, 21, 139–157. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.T.; Sobur, M.A.; Islam, M.S.; Ievy, S.; Hossain, M.J.; El Zowalaty, M.E.; Rahman, A.T.; Ashour, H.M. Zoonotic Diseases: Etiology, Impact, and Control. Microorganisms 2020, 8, 1405. [Google Scholar] [CrossRef] [PubMed]
- Schmeller, D.S.; Courchamp, F.; Killeen, G. Biodiversity loss, emerging pathogens and human health risks. Biodivers. Conserv. 2020, 29, 3095–3102. [Google Scholar] [CrossRef]
- Daszak, P.; Cunningham, A.A.; Hyatt, A.D. Anthropogenic environmental change and the emergence of infectious diseases in wildlife. Acta Trop. 2001, 78, 103–116. [Google Scholar] [CrossRef]
- Childs, J.E.; Richt, J.A.; Mackenzie, J.S. Introduction: Conceptualizing and partitioning the emergence process of zoonotic viruses from wildlife to humans. Curr. Top Microbiol. Immunol. 2007, 315, 1–31. [Google Scholar] [CrossRef]
- Calisher, C.H.; Childs, J.E.; Field, H.E.; Holmes, K.V.; Schountz, T. Bats: Important reservoir hosts of emerging viruses. Clin. Microbiol. Rev. 2006, 19, 531–545. [Google Scholar] [CrossRef]
- Serra-Cobo, J.; López-Roig, M. Bats and Emerging Infections: An Ecological and Virological Puzzle. Adv. Exp. Med. Biol. 2017, 972, 35–48. [Google Scholar] [CrossRef]
- Letko, M.; Seifert, S.N.; Olival, K.J.; Plowright, R.K.; Munster, V.J. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 2020, 18, 461–471. [Google Scholar] [CrossRef]
- Kohl, C.; Nitsche, A.; Kurth, A. Update on Potentially Zoonotic Viruses of European Bats. Vaccines 2021, 9, 690. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Victoria, J.G.; Wang, C.; Jones, M.; Fellers, G.M.; Kunz, T.H.; Delwart, E. Bat guano virome: Predominance of dietary viruses from insects and plants plus novel mammalian viruses. J. Virol. 2010, 84, 6955–6965. [Google Scholar] [CrossRef] [PubMed]
- Halpin, K.; Young, P.L.; Field, H.E.; Mackenzie, J.S. Isolation of Hendra virus from pteropid bats: A natural reservoir of Hendra virus. J. Gen. Virol. 2000, 81, 1927–1932. [Google Scholar] [CrossRef] [PubMed]
- Leroy, E.M.; Kumulungui, B.; Pourrut, X.; Rouquet, P.; Hassanin, A.; Yaba, P.; Délicat, A.; Paweska, J.T.; Gonzalez, J.P.; Swanepoel, R. Fruit bats as reservoirs of Ebola virus. Nature 2005, 438, 575–576. [Google Scholar] [CrossRef]
- Yob, J.M.; Field, H.; Rashdi, A.M.; Morrissy, C.; van der Heide, B.; Rota, P.; bin Adzhar, A.; White, J.; Daniels, P.; Jamaluddin, A.; et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001, 7, 439–441. [Google Scholar] [CrossRef]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H.; et al. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef]
- Wang, Q.; Qi, J.; Yuan, Y.; Xuan, Y.; Han, P.; Wan, Y.; Ji, W.; Li, Y.; Wu, Y.; Wang, J.; et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe 2014, 16, 328–337. [Google Scholar] [CrossRef]
- Memish, Z.A.; Mishra, N.; Olival, K.J.; Fagbo, S.F.; Kapoor, V.; Epstein, J.H.; Alhakeem, R.; Durosinloun, A.; Al Asmari, M.; Islam, A.; et al. Middle East respiratory syndrome coronavirus in bats, Saudi Arabia. Emerg. Infect. Dis. 2013, 19, 1819–1823. [Google Scholar] [CrossRef]
- Li, C.; Yang, Y.; Ren, L. Genetic evolution analysis of 2019 novel coronavirus and coronavirus from other species. Infect. Genet. Evol. 2020, 82, 104285. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Holmes, E.C.; Goldstein, S.A.; Rasmussen, A.L.; Robertson, D.L.; Crits-Christoph, A.; Wertheim, J.O.; Anthony, S.J.; Barclay, W.S.; Boni, M.F.; Doherty, P.C.; et al. The origins of SARS-CoV-2: A critical review. Cell 2021, 184, 4848–4856. [Google Scholar] [CrossRef] [PubMed]
- Li, L.L.; Wang, J.L.; Ma, X.H.; Sun, X.M.; Li, J.S.; Yang, X.F.; Shi, W.F.; Duan, Z.J. A novel SARS-CoV-2 related coronavirus with complex recombination isolated from bats in Yunnan province, China. Emerg. Microbes Infect. 2021, 10, 1683–1690. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Ji, J.; Chen, X.; Bi, Y.; Li, J.; Wang, Q.; Hu, T.; Song, H.; Zhao, R.; Chen, Y.; et al. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell 2021, 184, 4380–4391.e4314. [Google Scholar] [CrossRef]
- Plowright, R.K.; Field, H.E.; Smith, C.; Divljan, A.; Palmer, C.; Tabor, G.; Daszak, P.; Foley, J.E. Reproduction and nutritional stress are risk factors for Hendra virus infection in little red flying foxes (Pteropus scapulatus). Proc. Biol. Sci. 2008, 275, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Plowright, R.K.; Eby, P.; Hudson, P.J.; Smith, I.L.; Westcott, D.; Bryden, W.L.; Middleton, D.; Reid, P.A.; McFarlane, R.A.; Martin, G.; et al. Ecological dynamics of emerging bat virus spillover. Proc. Biol. Sci. 2015, 282, 20142124. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Corman, V.M.; Wegner, T.; Tateno, A.F.; Zerbinati, R.M.; Gloza-Rausch, F.; Seebens, A.; Müller, M.A.; Drosten, C. Amplification of emerging viruses in a bat colony. Emerg. Infect. Dis. 2011, 17, 449–456. [Google Scholar] [CrossRef]
- Fledermausschutz; Bat Conservation Switzerland. Available online: https://fledermausschutz.ch/ (accessed on 10 May 2022).
- UNEP/EUROBATS. Agreement on the Conservation of Populations of European Bats. Available online: https://www.eurobats.org/ (accessed on 10 May 2022).
- Shipley, R.; Wright, E.; Selden, D.; Wu, G.; Aegerter, J.; Fooks, A.R.; Banyard, A.C. Bats and Viruses: Emergence of Novel Lyssaviruses and Association of Bats with Viral Zoonoses in the EU. Trop. Med. Infect. Dis. 2019, 4, 31. [Google Scholar] [CrossRef]
- Dietz, C.; Nill, D.; Von Helversen, O. Bats of Britain, Europe and Northwest Africa; A & C Black Publishers Ltd.: London, UK, 2009. [Google Scholar]
- Dacheux, L.; Cervantes-Gonzalez, M.; Guigon, G.; Thiberge, J.M.; Vandenbogaert, M.; Maufrais, C.; Caro, V.; Bourhy, H. A preliminary study of viral metagenomics of French bat species in contact with humans: Identification of new mammalian viruses. PLoS ONE 2014, 9, e87194. [Google Scholar] [CrossRef]
- Zheng, X.Y.; Qiu, M.; Guan, W.J.; Li, J.M.; Chen, S.W.; Cheng, M.J.; Huo, S.T.; Chen, Z.; Wu, Y.; Jiang, L.N.; et al. Viral metagenomics of six bat species in close contact with humans in southern China. Arch. Virol. 2018, 163, 73–88. [Google Scholar] [CrossRef]
- Mishra, N.; Fagbo, S.F.; Alagaili, A.N.; Nitido, A.; Williams, S.H.; Ng, J.; Lee, B.; Durosinlorun, A.; Garcia, J.A.; Jain, K.; et al. A viral metagenomic survey identifies known and novel mammalian viruses in bats from Saudi Arabia. PLoS ONE 2019, 14, e0214227. [Google Scholar] [CrossRef] [Green Version]
- Wu, Z.; Yang, L.; Ren, X.; He, G.; Zhang, J.; Yang, J.; Qian, Z.; Dong, J.; Sun, L.; Zhu, Y.; et al. Deciphering the bat virome catalog to better understand the ecological diversity of bat viruses and the bat origin of emerging infectious diseases. ISME J. 2016, 10, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Geldenhuys, M.; Mortlock, M.; Weyer, J.; Bezuidt, O.; Seamark, E.C.J.; Kearney, T.; Gleasner, C.; Erkkila, T.H.; Cui, H.; Markotter, W. A metagenomic viral discovery approach identifies potential zoonotic and novel mammalian viruses in Neoromicia bats within South Africa. PLoS ONE 2018, 13, e0194527. [Google Scholar] [CrossRef]
- Li, Y.; Altan, E.; Reyes, G.; Halstead, B.; Deng, X.; Delwart, E. Virome of Bat Guano from Nine Northern California Roosts. J. Virol. 2021, 95, e01713-20. [Google Scholar] [CrossRef]
- Bolatti, E.M.; Viarengo, G.; Zorec, T.M.; Cerri, A.; Montani, M.E.; Hosnjak, L.; Casal, P.E.; Bortolotto, E.; Di Domenica, V.; Chouhy, D.; et al. Viral Metagenomic Data Analyses of Five New World Bat Species from Argentina: Identification of 35 Novel DNA Viruses. Microorganisms 2022, 10, 266. [Google Scholar] [CrossRef]
- Donaldson, E.F.; Haskew, A.N.; Gates, J.E.; Huynh, J.; Moore, C.J.; Frieman, M.B. Metagenomic analysis of the viromes of three North American bat species: Viral diversity among different bat species that share a common habitat. J. Virol. 2010, 84, 13004–13018. [Google Scholar] [CrossRef] [PubMed]
- Hardmeier, I.; Aeberhard, N.; Qi, W.; Schoenbaechler, K.; Kraettli, H.; Hatt, J.M.; Fraefel, C.; Kubacki, J. Metagenomic analysis of fecal and tissue samples from 18 endemic bat species in Switzerland revealed a diverse virus composition including potentially zoonotic viruses. PLoS ONE 2021, 16, e0252534. [Google Scholar] [CrossRef]
- Šimić, I.; Zorec, T.M.; Lojkić, I.; Krešić, N.; Poljak, M.; Cliquet, F.; Picard-Meyer, E.; Wasniewski, M.; Zrnčić, V.; Ćukušić, A.; et al. Viral Metagenomic Profiling of Croatian Bat Population Reveals Sample and Habitat Dependent Diversity. Viruses 2020, 12, 891. [Google Scholar] [CrossRef]
- Mendenhall, I.H.; Wen, D.L.H.; Jayakumar, J.; Gunalan, V.; Wang, L.; Mauer-Stroh, S.; Su, Y.C.F.; Smith, G.J.D. Diversity and Evolution of Viral Pathogen Community in Cave Nectar Bats. Viruses 2019, 11, 250. [Google Scholar] [CrossRef]
- Obameso, J.O.; Li, H.; Jia, H.; Han, M.; Zhu, S.; Huang, C.; Zhao, Y.; Zhao, M.; Bai, Y.; Yuan, F.; et al. The persistent prevalence and evolution of cross-family recombinant coronavirus GCCDC1 among a bat population: A two-year follow-up. Sci. China Life Sci. 2017, 60, 1357–1363. [Google Scholar] [CrossRef]
- Pauly, M.; Pir, J.B.; Loesch, C.; Sausy, A.; Snoeck, C.J.; Hübschen, J.M.; Muller, C.P. Novel Alphacoronaviruses and Paramyxoviruses Cocirculate with Type 1 and Severe Acute Respiratory System (SARS)-Related Betacoronaviruses in Synanthropic Bats of Luxembourg. Appl. Environ. Microbiol. 2017, 83, e01326-17. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.Y.; Wang, N.; Zhang, W.; Hu, B.; Li, B.; Zhang, Y.Z.; Zhou, J.H.; Luo, C.M.; Yang, X.L.; Wu, L.J.; et al. Coexistence of multiple coronaviruses in several bat colonies in an abandoned mineshaft. Virol. Sin. 2016, 31, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Kubacki, J.; Fraefel, C.; Bachofen, C. Implementation of next-generation sequencing for virus identification in veterinary diagnostic laboratories. J. Vet. Diagn. Investig. 2020, 33, 235–247. [Google Scholar] [CrossRef]
- Kubacki, J.; Flacio, E.; Qi, W.; Guidi, V.; Tonolla, M.; Fraefel, C. Viral Metagenomic Analysis of Aedes albopictus Mosquitos from Southern Switzerland. Viruses 2020, 12, 929. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. metaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Liu, B.; Yang, J.; Jin, Q. DBatVir: The database of bat-associated viruses. Database 2014, 2014, bau021. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Garrison, E.; Marth, G. Haplotype-Based Variant Detection from Short-Read Sequencing. arXiv 2012, arXiv:1207.3907. [Google Scholar]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Subgroup, G.P.D.P. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Salmier, A.; Tirera, S.; de Thoisy, B.; Franc, A.; Darcissac, E.; Donato, D.; Bouchier, C.; Lacoste, V.; Lavergne, A. Virome analysis of two sympatric bat species (Desmodus rotundus and Molossus molossus) in French Guiana. PLoS ONE 2017, 12, e0186943. [Google Scholar] [CrossRef]
- Hu, D.; Zhu, C.; Wang, Y.; Ai, L.; Yang, L.; Ye, F.; Ding, C.; Chen, J.; He, B.; Zhu, J.; et al. Virome analysis for identification of novel mammalian viruses in bats from Southeast China. Sci. Rep. 2017, 7, 10917. [Google Scholar] [CrossRef]
- Shi, S.L.; Xia, R.X. Codon Usage in the Iflaviridae Family Is Not Diverse Though the Family Members Are Isolated from Diverse Host Taxa. Viruses 2019, 11, 1087. [Google Scholar] [CrossRef]
- Genersch, E.; Aubert, M. Emerging and re-emerging viruses of the honey bee (Apis mellifera L.). Vet. Res. 2010, 41, 54. [Google Scholar] [CrossRef]
- Wang, F.; Yuan, B.; Xiao, S.; Zhang, J.; Jia, W.; Fang, Q.; Song, Q.; Ye, G. Diverse RNA Viruses Discovered in Three Parasitoid Wasps of the Rice Weevil. mSphere 2021, 6, e00331-21. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, Y.C.; Wang, Z.G.; Gu, Q.Y.; Niu, J.Z.; Wang, J.J. The Diversity of Viral Community in Invasive Fruit Flies (Bactrocera and Zeugodacus) Revealed by Meta-transcriptomics. Microb. Ecol. 2022, 83, 739–752. [Google Scholar] [CrossRef]
- Langat, S.K.; Eyase, F.; Bulimo, W.; Lutomiah, J.; Oyola, S.O.; Imbuga, M.; Sang, R. Profiling of RNA Viruses in Biting Midges. mSphere 2021, 6, e0055121. [Google Scholar] [CrossRef]
- Jia, W.; Wang, F.; Li, J.; Chang, X.; Yang, Y.; Yao, H.; Bao, Y.; Song, Q.; Ye, G. A Novel Iflavirus Was Discovered in Green Rice Leafhopper. Front. Microbiol. 2020, 11, 621141. [Google Scholar] [CrossRef] [PubMed]
- Valles, S.M.; Chen, Y.; Firth, A.E.; Guérin, D.M.A.; Hashimoto, Y.; Herrero, S.; de Miranda, J.R.; Ryabov, E.; Ictv Report Consortium. ICTV Virus Taxonomy Profile: Iflaviridae. J. Gen. Virol. 2017, 98, 527–528. [Google Scholar] [CrossRef] [PubMed]
- ICTV Virus Taxonomy Profile: Subfamily Densovirinae. Available online: https://talk.ictvonline.org/ictv-reports/ictv_online_report/ssdna-viruses/w/parvoviridae/1047/subfamily-densovirinae (accessed on 8 June 2022).
- Ge, X.; Li, Y.; Yang, X.; Zhang, H.; Zhou, P.; Zhang, Y.; Shi, Z. Metagenomic analysis of viruses from bat fecal samples reveals many novel viruses in insectivorous bats in China. J. Virol. 2012, 86, 4620–4630. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, M.; Mühldorfer, K.; Speck, S.; Wibbelt, G.; Kurth, A. New adenovirus in bats, Germany. Emerg. Infect. Dis. 2009, 15, 2052–2055. [Google Scholar] [CrossRef]
- Waruhiu, C.; Ommeh, S.; Obanda, V.; Agwanda, B.; Gakuya, F.; Ge, X.Y.; Yang, X.L.; Wu, L.J.; Zohaib, A.; Hu, B.; et al. Molecular detection of viruses in Kenyan bats and discovery of novel astroviruses, caliciviruses and rotaviruses. Virol. Sin. 2017, 32, 101–114. [Google Scholar] [CrossRef]
- Garrison, A.R.; Alkhovsky Альхoвский Сергей Владимирoвич, S.V.; Avšič-Županc, T.; Bente, D.A.; Bergeron, É.; Burt, F.; Di Paola, N.; Ergünay, K.; Hewson, R.; Kuhn, J.H.; et al. ICTV Virus Taxonomy Profile Nairoviridae. J. Gen. Virol. 2020, 101, 798–799. [Google Scholar] [CrossRef]
- Brinkmann, A.; Kohl, C.; Radonić, A.; Dabrowski, P.W.; Mühldorfer, K.; Nitsche, A.; Wibbelt, G.; Kurth, A. First detection of bat-borne Issyk-Kul virus in Europe. Sci. Rep. 2020, 10, 22384. [Google Scholar] [CrossRef]
- Tao, Y.; Shi, M.; Conrardy, C.; Kuzmin, I.V.; Recuenco, S.; Agwanda, B.; Alvarez, D.A.; Ellison, J.A.; Gilbert, A.T.; Moran, D.; et al. Discovery of diverse polyomaviruses in bats and the evolutionary history of the Polyomaviridae. J. Gen. Virol. 2013, 94, 738–748. [Google Scholar] [CrossRef]
- Fagrouch, Z.; Sarwari, R.; Lavergne, A.; Delaval, M.; de Thoisy, B.; Lacoste, V.; Verschoor, E.J. Novel polyomaviruses in South American bats and their relationship to other members of the family Polyomaviridae. J. Gen. Virol. 2012, 93, 2652–2657. [Google Scholar] [CrossRef]
- Misra, V.; Dumonceaux, T.; Dubois, J.; Willis, C.; Nadin-Davis, S.; Severini, A.; Wandeler, A.; Lindsay, R.; Artsob, H. Detection of polyoma and corona viruses in bats of Canada. J. Gen. Virol. 2009, 90, 2015–2022. [Google Scholar] [CrossRef]
- Vidovszky, M.Z.; Tan, Z.; Carr, M.J.; Boldogh, S.; Harrach, B.; Gonzalez, G. Bat-borne polyomaviruses in Europe reveal an evolutionary history of intrahost divergence with horseshoe bats distributed across the African and Eurasian continents. J. Gen. Virol. 2020, 101, 1119–1130. [Google Scholar] [CrossRef] [PubMed]
- Satterfield, D.A.; Marra, P.P.; Sillett, T.S.; Altizer, S. Responses of migratory species and their pathogens to supplemental feeding. Philos. Trans. R Soc. Lond. B Biol. Sci. 2018, 373, 20170094. [Google Scholar] [CrossRef] [PubMed]
- Lelli, D.; Papetti, A.; Sabelli, C.; Rosti, E.; Moreno, A.; Boniotti, M.B. Detection of coronaviruses in bats of various species in Italy. Viruses 2013, 5, 2679–2689. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Corman, V.M.; Drosten, C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antivir. Res. 2014, 101, 45–56. [Google Scholar] [CrossRef]
- Drexler, J.F.; Gloza-Rausch, F.; Glende, J.; Corman, V.M.; Muth, D.; Goettsche, M.; Seebens, A.; Niedrig, M.; Pfefferle, S.; Yordanov, S.; et al. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J. Virol. 2010, 84, 11336–11349. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, F.; Edenborough, K.M.; Toffoli, R.; Culasso, P.; Zoppi, S.; Dondo, A.; Robetto, S.; Rosati, S.; Lander, A.; Kurth, A.; et al. Coronavirus and paramyxovirus in bats from Northwest Italy. BMC Vet. Res. 2017, 13, 396. [Google Scholar] [CrossRef] [PubMed]
- Rihtaric, D.; Hostnik, P.; Steyer, A.; Grom, J.; Toplak, I. Identification of SARS-like coronaviruses in horseshoe bats (Rhinolophus hipposideros) in Slovenia. Arch. Virol. 2010, 155, 507–514. [Google Scholar] [CrossRef]
- Alkhovsky, S.; Lenshin, S.; Romashin, A.; Vishnevskaya, T.; Vyshemirsky, O.; Bulycheva, Y.; Lvov, D.; Gitelman, A. SARS-like Coronaviruses in Horseshoe Bats. Viruses 2022, 14, 113. [Google Scholar] [CrossRef]
- Annan, A.; Baldwin, H.J.; Corman, V.M.; Klose, S.M.; Owusu, M.; Nkrumah, E.E.; Badu, E.K.; Anti, P.; Agbenyega, O.; Meyer, B.; et al. Human betacoronavirus 2c EMC/2012-related viruses in bats, Ghana and Europe. Emerg. Infect. Dis. 2013, 19, 456–459. [Google Scholar] [CrossRef]
- Crook, J.M.; Murphy, I.; Carter, D.P.; Pullan, S.T.; Carroll, M.; Vipond, R.; Cunningham, A.A.; Bell, D. Metagenomic identification of a new sarbecovirus from horseshoe bats in Europe. Sci. Rep. 2021, 11, 14723. [Google Scholar] [CrossRef]
- Ar Gouilh, M.; Puechmaille, S.J.; Diancourt, L.; Vandenbogaert, M.; Serra-Cobo, J.; Lopez Roïg, M.; Brown, P.; Moutou, F.; Caro, V.; Vabret, A.; et al. SARS-CoV related Betacoronavirus and diverse Alphacoronavirus members found in western old-world. Virology 2018, 517, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Al-Salihi, K.A.; Khalaf, J.M. The emerging SARS-CoV, MERS-CoV, and SARS-CoV-2: An insight into the viruses zoonotic aspects. Vet. World 2021, 14, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Mohd, H.A.; Al-Tawfiq, J.A.; Memish, Z.A. Middle East Respiratory Syndrome Coronavirus (MERS-CoV) origin and animal reservoir. Virol. J. 2016, 13, 87. [Google Scholar] [CrossRef] [PubMed]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef] [PubMed]
- Anthony, S.J.; Gilardi, K.; Menachery, V.D.; Goldstein, T.; Ssebide, B.; Mbabazi, R.; Navarrete-Macias, I.; Liang, E.; Wells, H.; Hicks, A.; et al. Further Evidence for Bats as the Evolutionary Source of Middle East Respiratory Syndrome Coronavirus. mBio 2017, 8, e00373-17. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A.; et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef]
- Luo, C.M.; Wang, N.; Yang, X.L.; Liu, H.Z.; Zhang, W.; Li, B.; Hu, B.; Peng, C.; Geng, Q.B.; Zhu, G.J.; et al. Discovery of Novel Bat Coronaviruses in South China That Use the Same Receptor as Middle East Respiratory Syndrome Coronavirus. J. Virol. 2018, 92, e00116-18. [Google Scholar] [CrossRef]
- Lau, S.K.P.; Fan, R.Y.Y.; Zhu, L.; Li, K.S.M.; Wong, A.C.P.; Luk, H.K.H.; Wong, E.Y.M.; Lam, C.S.F.; Lo, G.C.S.; Fung, J.; et al. Isolation of MERS-related coronavirus from lesser bamboo bats that uses DPP4 and infects human-DPP4-transgenic mice. Nat. Commun. 2021, 12, 216. [Google Scholar] [CrossRef]
- Moya, A.; Holmes, E.C.; González-Candelas, F. The population genetics and evolutionary epidemiology of RNA viruses. Nat. Rev. Microbiol. 2004, 2, 279–288. [Google Scholar] [CrossRef]
- Woolhouse, M.E.; Gowtage-Sequeria, S. Host range and emerging and reemerging pathogens. Emerg. Infect. Dis. 2005, 11, 1842–1847. [Google Scholar] [CrossRef]
- Lau, S.K.; Woo, P.C.; Li, K.S.; Huang, Y.; Tsoi, H.W.; Wong, B.H.; Wong, S.S.; Leung, S.Y.; Chan, K.H.; Yuen, K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 2005, 102, 14040–14045. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Xing, N.; Meng, K.; Fu, B.; Xue, W.; Dong, P.; Tang, W.; Xiao, Y.; Liu, G.; Luo, H.; et al. Nucleocapsid mutations R203K/G204R increase the infectivity, fitness, and virulence of SARS-CoV-2. Cell Host Microbe 2021, 29, 1788–1801.e1786. [Google Scholar] [CrossRef] [PubMed]
- Van Cleemput, J.; van Snippenberg, W.; Lambrechts, L.; Dendooven, A.; D’Onofrio, V.; Couck, L.; Trypsteen, W.; Vanrusselt, J.; Theuns, S.; Vereecke, N.; et al. Organ-specific genome diversity of replication-competent SARS-CoV-2. Nat. Commun. 2021, 12, 6612. [Google Scholar] [CrossRef] [PubMed]
- Rantam, F.A.; Prakoeswa, C.R.S.; Tinduh, D.; Nugraha, J.; Susilowati, H.; Wijaya, A.Y.; Puspaningsih, N.N.T.; Puspitasari, D.; Husada, D.; Kurniati, N.D.; et al. Characterization of SARS-CoV-2 East Java isolate, Indonesia. F1000Res 2021, 10, 480. [Google Scholar] [CrossRef]
- Molina-Mora, J.A.; Cordero-Laurent, E.; Godínez, A.; Calderón-Osorno, M.; Brenes, H.; Soto-Garita, C.; Pérez-Corrales, C.; Drexler, J.F.; Moreira-Soto, A.; Corrales-Aguilar, E.; et al. SARS-CoV-2 genomic surveillance in Costa Rica: Evidence of a divergent population and an increased detection of a spike T1117I mutation. Infect. Genet. Evol. 2021, 92, 104872. [Google Scholar] [CrossRef]
- Kosuge, M.; Furusawa-Nishii, E.; Ito, K.; Saito, Y.; Ogasawara, K. Point mutation bias in SARS-CoV-2 variants results in increased ability to stimulate inflammatory responses. Sci. Rep. 2020, 10, 17766. [Google Scholar] [CrossRef]
- Liu, Z.; Zheng, H.; Lin, H.; Li, M.; Yuan, R.; Peng, J.; Xiong, Q.; Sun, J.; Li, B.; Wu, J.; et al. Identification of Common Deletions in the Spike Protein of Severe Acute Respiratory Syndrome Coronavirus 2. J. Virol. 2020, 94, e00790-20. [Google Scholar] [CrossRef]
- Gong, Y.N.; Tsao, K.C.; Hsiao, M.J.; Huang, C.G.; Huang, P.N.; Huang, P.W.; Lee, K.M.; Liu, Y.C.; Yang, S.L.; Kuo, R.L.; et al. SARS-CoV-2 genomic surveillance in Taiwan revealed novel ORF8-deletion mutant and clade possibly associated with infections in Middle East. Emerg. Microbes Infect. 2020, 9, 1457–1466. [Google Scholar] [CrossRef]
- Graepel, K.W.; Lu, X.; Case, J.B.; Sexton, N.R.; Smith, E.C.; Denison, M.R. Proofreading-Deficient Coronaviruses Adapt for Increased Fitness over Long-Term Passage without Reversion of Exoribonuclease-Inactivating Mutations. mBio 2017, 8, e01503-17. [Google Scholar] [CrossRef]
- Duffy, S.; Shackelton, L.A.; Holmes, E.C. Rates of evolutionary change in viruses: Patterns and determinants. Nat. Rev. Genet. 2008, 9, 267–276. [Google Scholar] [CrossRef]
- Woo, P.C.; Huang, Y.; Lau, S.K.; Yuen, K.Y. Coronavirus genomics and bioinformatics analysis. Viruses 2010, 2, 1804–1820. [Google Scholar] [CrossRef]
- Tay, J.H.; Porter, A.F.; Wirth, W.; Duchene, S. The Emergence of SARS-CoV-2 Variants of Concern Is Driven by Acceleration of the Substitution Rate. Mol. Biol. Evol. 2022, 39, msac013. [Google Scholar] [CrossRef] [PubMed]
- Sahin, E.; Bozdayi, G.; Yigit, S.; Muftah, H.; Dizbay, M.; Tunccan, O.G.; Fidan, I.; Caglar, K. Genomic characterization of SARS-CoV-2 isolates from patients in Turkey reveals the presence of novel mutations in spike and nsp12 proteins. J. Med. Virol. 2021, 93, 6016–6026. [Google Scholar] [CrossRef] [PubMed]
- Das, J.K.; Roy, S. A study on non-synonymous mutational patterns in structural proteins of SARS-CoV-2. Genome 2021, 64, 665–678. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.P.; Lybrand, T.P.; Gilbert, P.B.; Hawn, T.R.; Schiffer, J.T.; Stamatatos, L.; Payne, T.H.; Carpp, L.N.; Geraghty, D.E.; Jerome, K.R. Tracking SARS-CoV-2 Spike Protein Mutations in the United States (January 2020–March 2021) Using a Statistical Learning Strategy. Viruses 2021, 14, 9. [Google Scholar] [CrossRef]
- Rahbar, M.R.; Jahangiri, A.; Khalili, S.; Zarei, M.; Mehrabani-Zeinabad, K.; Khalesi, B.; Pourzardosht, N.; Hessami, A.; Nezafat, N.; Sadraei, S.; et al. Hotspots for mutations in the SARS-CoV-2 spike glycoprotein: A correspondence analysis. Sci. Rep. 2021, 11, 23622. [Google Scholar] [CrossRef]
- Graham, R.L.; Baric, R.S. Recombination, reservoirs, and the modular spike: Mechanisms of coronavirus cross-species transmission. J. Virol. 2010, 84, 3134–3146. [Google Scholar] [CrossRef]
- Paul, D.; Pyne, N.; Paul, S. Mutation profile of SARS-CoV-2 spike protein and identification of potential multiple epitopes within spike protein for vaccine development against SARS-CoV-2. Virusdisease 2021, 32, 703–726. [Google Scholar] [CrossRef]
- Rouchka, E.C.; Chariker, J.H.; Chung, D. Variant analysis of 1,040 SARS-CoV-2 genomes. PLoS ONE 2020, 15, e0241535. [Google Scholar] [CrossRef]
- Kemp, S.A.; Collier, D.A.; Datir, R.P.; Ferreira, I.A.T.M.; Gayed, S.; Jahun, A.; Hosmillo, M.; Rees-Spear, C.; Mlcochova, P.; Lumb, I.U.; et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature 2021, 592, 277–282. [Google Scholar] [CrossRef]
- O’Shea, T.J.; Cryan, P.M.; Cunningham, A.A.; Fooks, A.R.; Hayman, D.T.; Luis, A.D.; Peel, A.J.; Plowright, R.K.; Wood, J.L. Bat flight and zoonotic viruses. Emerg. Infect. Dis. 2014, 20, 741–745. [Google Scholar] [CrossRef] [PubMed]
- Baker, M.L.; Schountz, T.; Wang, L.F. Antiviral immune responses of bats: A review. Zoonoses Public Health 2013, 60, 104–116. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, G.S.; Adams, D.M. Recurrent evolution of extreme longevity in bats. Biol. Lett. 2019, 15, 20180860. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.R.; Behan, W.M. High mutation rate in the NS1 gene of parvovirus B19 DNA amplified from skeletal muscle of a case of mixed connective tissue disease. Rheumatology 2002, 41, 833–834. [Google Scholar] [CrossRef] [PubMed]
- López-Bueno, A.; Villarreal, L.P.; Almendral, J.M. Parvovirus variation for disease: A difference with RNA viruses? Curr Top Microbiol. Immunol. 2006, 299, 349–370. [Google Scholar] [CrossRef]
- Shackelton, L.A.; Parrish, C.R.; Truyen, U.; Holmes, E.C. High rate of viral evolution associated with the emergence of carnivore parvovirus. Proc. Natl. Acad. Sci. USA 2005, 102, 379–384. [Google Scholar] [CrossRef]
- Abdelrahman, D.; Al-Sadeq, D.W.; Smatti, M.K.; Taleb, S.A.; AbuOdeh, R.O.; Al-Absi, E.S.; Al-Thani, A.A.; Coyle, P.V.; Al-Dewik, N.; Qahtani, A.A.A.; et al. Prevalence and Phylogenetic Analysis of Parvovirus (B19V) among Blood Donors with Different Nationalities Residing in Qatar. Viruses 2021, 13, 540. [Google Scholar] [CrossRef]
- Franzo, G.; He, W.; Correa-Fiz, F.; Li, G.; Legnardi, M.; Su, S.; Segalés, J. A Shift in Porcine Circovirus 3 (PCV-3) History Paradigm: Phylodynamic Analyses Reveal an Ancient Origin and Prolonged Undetected Circulation in the Worldwide Swine Population. Adv. Sci. 2019, 6, 1901004. [Google Scholar] [CrossRef]
- Dei Giudici, S.; Franzoni, G.; Bonelli, P.; Angioi, P.P.; Zinellu, S.; Deriu, V.; Carta, T.; Sechi, A.M.; Salis, F.; Balzano, F.; et al. Genetic Characterization of Porcine Circovirus 3 Strains Circulating in Sardinian Pigs and Wild Boars. Pathogens 2020, 9, 344. [Google Scholar] [CrossRef]
- Umemura, T.; Tanaka, Y.; Kiyosawa, K.; Alter, H.J.; Shih, J.W. Observation of positive selection within hypervariable regions of a newly identified DNA virus (SEN virus)(1). FEBS Lett. 2002, 510, 171–174. [Google Scholar] [CrossRef] [Green Version]
- Sarker, S.; Patterson, E.I.; Peters, A.; Baker, G.B.; Forwood, J.K.; Ghorashi, S.A.; Holdsworth, M.; Baker, R.; Murray, N.; Raidal, S.R. Mutability dynamics of an emergent single stranded DNA virus in a naïve host. PLoS ONE 2014, 9, e85370. [Google Scholar] [CrossRef]
- Kundu, S.; Faulkes, C.G.; Greenwood, A.G.; Jones, C.G.; Kaiser, P.; Lyne, O.D.; Black, S.A.; Chowrimootoo, A.; Groombridge, J.J. Tracking viral evolution during a disease outbreak: The rapid and complete selective sweep of a circovirus in the endangered Echo parakeet. J. Virol. 2012, 86, 5221–5229. [Google Scholar] [CrossRef] [PubMed]
- Johne, R.; Fernández-de-Luco, D.; Höfle, U.; Müller, H. Genome of a novel circovirus of starlings, amplified by multiply primed rolling-circle amplification. J. Gen. Virol. 2006, 87, 1189–1195. [Google Scholar] [CrossRef] [PubMed]



| Colony | Virus | Gene | Length | nt-Substitutions s/ns |
|---|---|---|---|---|
| AG | Bat circovirus | Rep | 920nt | 2/0 |
| Cap | 845nt | 5/0 | ||
| Bat-associated circovirus | Rep | 920nt | 10/3 | |
| Cap | 779nt | 14/0 | ||
| Pipistrellus pipistrellus polyomavirus | LTAg | 2’512nt | 1/0 | |
| GR | AAV2 | Rep68 | 1’609nt | 0/2 |
| Rep78 | 1’865nt | 0/2 | ||
| Rep20 | 937nt | 0/2 | ||
| Rep52 | 1’193nt | 0/2 | ||
| Starling circovirus | Cap | 830nt | 1/0 | |
| LU | Cyclovirus | Rep | 830nt | 0/6 |
| Circovirus | Rep | 830nt | 3/3 | |
| Dependoparvovirus | Cap | 494nt | 3/1 | |
| Bat AAV | Cap | 2’175nt | 2/2 | |
| Murine-associated porcine bocavirus | NP1 | 599nt | 1/0 | |
| VP1 | 1’850nt | 3/0 |
| Colony | Virus | Gene | Length | nt-Substitutions s/ns | Novel |
|---|---|---|---|---|---|
| AG | MERS-related CoV | ORF1ab | 21’322nt | 172/14 | 14 |
| S | 4’049nt | 9/3 | 3 | ||
| ORF4a | 284nt | 0/1 | 1 | ||
| ORF4b | 698nt | 1/1 | 1 | ||
| ORF5 | 683nt | 3/1 | 1 | ||
| E | 248nt | 2/0 | 0 | ||
| M | 656nt | 8/0 | 0 | ||
| N | 1’301nt | 8/2 | 2 | ||
| ORF8b | 581nt | 1/4 | 3 | ||
| GR | Bat rotavirus | NSP3 | 942nt | 1/0 | 0 |
| VP4 | 2’331nt | 1/0 | 0 | ||
| Mammalian orthoreovirus | S1 | 1’417nt | 0/1 | 1 | |
| L3 | 3’901nt | 1/0 | 0 | ||
| LU | Alpha CoV | ORF1ab | 20’230nt | 184/35 | 20 |
| S | 4’094nt | 75/24 | 23 | ||
| ORF3 | 674nt | 5/7 | 2 | ||
| E | 230nt | 1/0 | 0 | ||
| N | 1’259nt | 0/2 | 2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wiederkehr, M.A.; Qi, W.; Schoenbaechler, K.; Fraefel, C.; Kubacki, J. Virus Diversity, Abundance, and Evolution in Three Different Bat Colonies in Switzerland. Viruses 2022, 14, 1911. https://doi.org/10.3390/v14091911
Wiederkehr MA, Qi W, Schoenbaechler K, Fraefel C, Kubacki J. Virus Diversity, Abundance, and Evolution in Three Different Bat Colonies in Switzerland. Viruses. 2022; 14(9):1911. https://doi.org/10.3390/v14091911
Chicago/Turabian StyleWiederkehr, Myriam Anja, Weihong Qi, Katja Schoenbaechler, Cornel Fraefel, and Jakub Kubacki. 2022. "Virus Diversity, Abundance, and Evolution in Three Different Bat Colonies in Switzerland" Viruses 14, no. 9: 1911. https://doi.org/10.3390/v14091911






