Cross-Reactivity of SARS-CoV-2 Nucleocapsid-Binding Antibodies and Its Implication for COVID-19 Serology Tests
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Viruses and Proteins
- HCoV-19/Russia/StPetersburg-3524/2020 (B.1 Lineage, Wuhan);
- HCoV-19/Russia/SPE-RII-27029S/2021 (B.1.351 Lineage, Beta);
- HCoV-19/Japan/TY7-503/2021 (P.1 Lineage, Gamma);
- HCoV-19/Russia/SPE-RII-32759S/2021 (B.1.617.2 Lineage, Delta);
- HCoV-19/Russia/SPE-RII-6243V1/2021 (B.1.1.529 Lineage, Omicron).
2.3. Generation of Recombinant N Proteins
2.4. Assessment of Virus-Specific Antibody in Human Serum Samples
2.5. Statistical Analysis
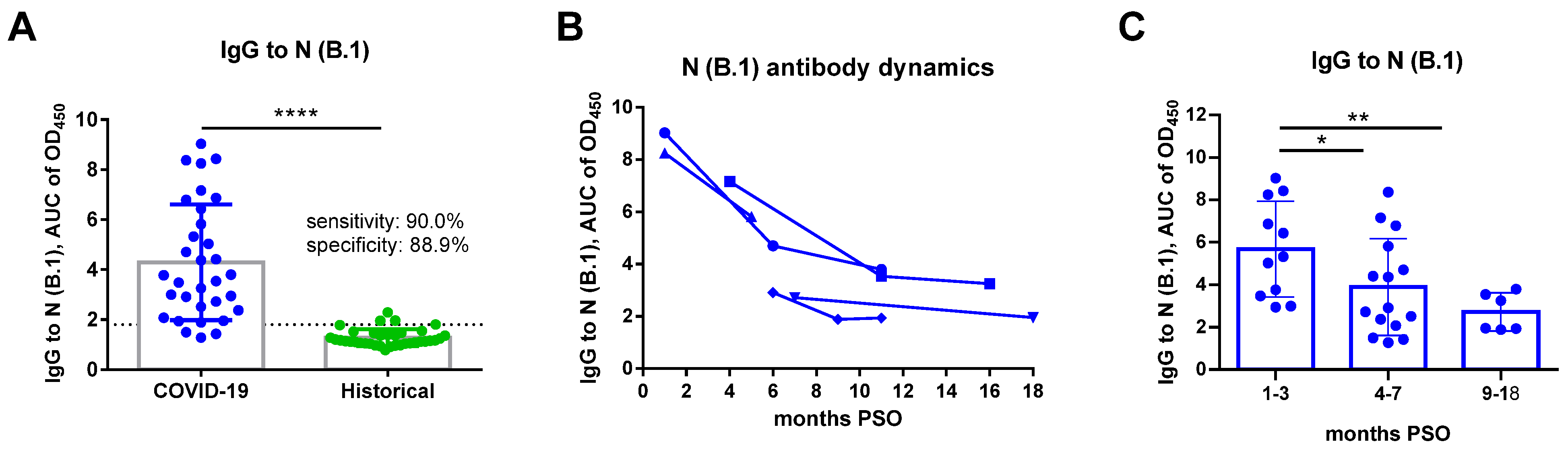
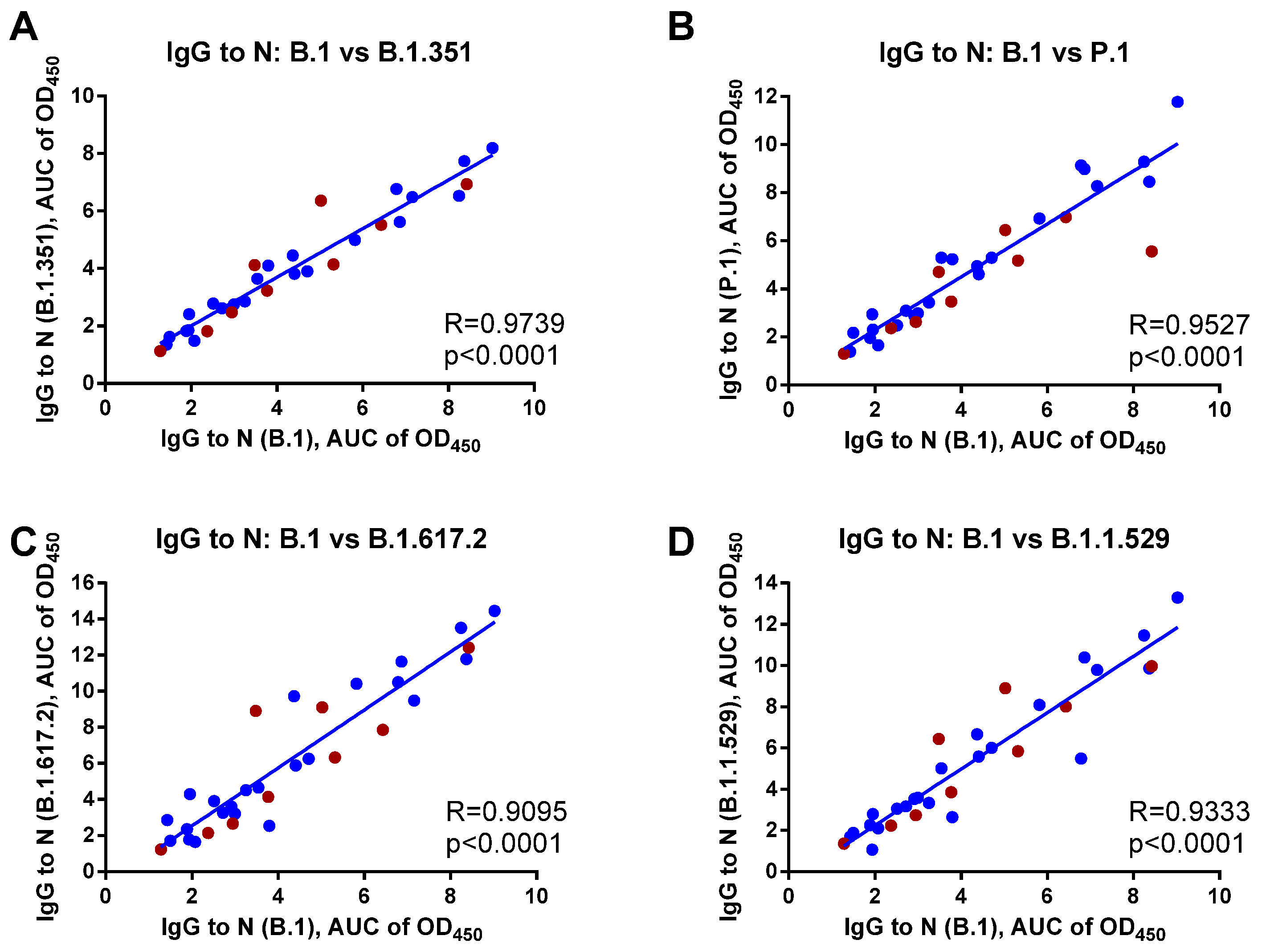
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, C.; Horby, P.W.; Hayden, F.G.; Gao, G.F. A novel coronavirus outbreak of global health concern. Lancet 2020, 395, 470–473. [Google Scholar] [CrossRef]
- Anonymous. Worldometer of COVID-19 Coronavirus Pandemic. Available online: https://www.worldometers.info/coronavirus/ (accessed on 13 June 2022).
- Safiabadi Tali, S.H.; LeBlanc, J.J.; Sadiq, Z.; Oyewunmi, O.D.; Camargo, C.; Nikpour, B.; Armanfard, N.; Sagan, S.M.; Jahanshahi-Anbuhi, S. Tools and Techniques for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2)/COVID-19 Detection. Clin. Microbiol. Rev. 2021, 34, e00228-20. [Google Scholar] [CrossRef] [PubMed]
- Peeling, R.W.; Wedderburn, C.J.; Garcia, P.J.; Boeras, D.; Fongwen, N.; Nkengasong, J.; Sall, A.; Tanuri, A.; Heymann, D.L. Serology testing in the COVID-19 pandemic response. Lancet. Infect. Dis. 2020, 20, e245–e249. [Google Scholar] [CrossRef]
- Fernandez-Barat, L.; Lopez-Aladid, R.; Torres, A. The value of serology testing to manage SARS-CoV-2 infections. Eur. Respir. J. 2020, 56, 2002411. [Google Scholar] [CrossRef] [PubMed]
- Upreti, S.; Samant, M. A Review on Immunological Responses to SARS-CoV-2 and Various COVID-19 Vaccine Regimens. Pharm. Res. 2022. [Google Scholar] [CrossRef]
- Shang, J.; Wan, Y.; Luo, C.; Ye, G.; Geng, Q.; Auerbach, A.; Li, F. Cell entry mechanisms of SARS-CoV-2. Proc. Natl. Acad. Sci. USA 2020, 117, 11727–11734. [Google Scholar] [CrossRef]
- Rockstroh, A.; Wolf, J.; Fertey, J.; Kalbitz, S.; Schroth, S.; Lubbert, C.; Ulbert, S.; Borte, S. Correlation of humoral immune responses to different SARS-CoV-2 antigens with virus neutralizing antibodies and symptomatic severity in a German COVID-19 cohort. Emerg. Microbes Infect. 2021, 10, 774–781. [Google Scholar] [CrossRef]
- McAndrews, K.M.; Dowlatshahi, D.P.; Dai, J.; Becker, L.M.; Hensel, J.; Snowden, L.M.; Leveille, J.M.; Brunner, M.R.; Holden, K.W.; Hopkins, N.S.; et al. Heterogeneous antibodies against SARS-CoV-2 spike receptor binding domain and nucleocapsid with implications for COVID-19 immunity. JCI Insight 2020, 5, e142386. [Google Scholar] [CrossRef]
- Perkmann, T.; Perkmann-Nagele, N.; Koller, T.; Mucher, P.; Radakovics, A.; Marculescu, R.; Wolzt, M.; Wagner, O.F.; Binder, C.J.; Haslacher, H. Anti-Spike Protein Assays to Determine SARS-CoV-2 Antibody Levels: A Head-to-Head Comparison of Five Quantitative Assays. Microbiol. Spectr. 2021, 9, e0024721. [Google Scholar] [CrossRef]
- Evolutionary insight into the emergence of SARS-CoV-2 variants of concern. Nat. Med. 2022, 28, 1357–1358. [CrossRef]
- Isakova-Sivak, I.; Stukova, M.; Erofeeva, M.; Naykhin, A.; Donina, S.; Petukhova, G.; Kuznetsova, V.; Kiseleva, I.; Smolonogina, T.; Dubrovina, I.; et al. H2N2 live attenuated influenza vaccine is safe and immunogenic for healthy adult volunteers. Hum. Vaccines Immunother. 2015, 11, 970–982. [Google Scholar] [CrossRef] [PubMed]
- Rudenko, L.; Kiseleva, I.; Stukova, M.; Erofeeva, M.; Naykhin, A.; Donina, S.; Larionova, N.; Pisareva, M.; Krivitskaya, V.; Flores, J.; et al. Clinical testing of pre-pandemic live attenuated A/H5N2 influenza candidate vaccine in adult volunteers: Results from a placebo-controlled, randomized double-blind phase I study. Vaccine 2015, 33, 5110–5117. [Google Scholar] [CrossRef] [PubMed]
- Matyushenko, V.; Isakova-Sivak, I.; Kudryavtsev, I.; Goshina, A.; Chistyakova, A.; Stepanova, E.; Prokopenko, P.; Sychev, I.; Rudenko, L. Detection of IFNgamma-Secreting CD4(+) and CD8(+) Memory T Cells in COVID-19 Convalescents after Stimulation of Peripheral Blood Mononuclear Cells with Live SARS-CoV-2. Viruses 2021, 13, 1490. [Google Scholar] [CrossRef]
- Prokofyev, A.; Gershovich, P.; Strelkova, A.; Spirina, N.; Kondinskaya, D.; Yakovlev, P.; Morozov, D. AAV5-Based Vaccine for Induction of Specific Immunity to SARS-CoV-2 Virus and/or Prevention of SARS-CoV-2-Induced Coronavirus Infection. Patent RU 2761879 C1, 13 December 2021. [Google Scholar]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed]
- Wong, J.W.; Albright, R.L.; Wang, N.-H.L. Immobilized Metal Ion Affinity Chromatography (IMAC) Chemistry and Bioseparation Applications. Sep. Purif. Methods 1991, 20, 49–106. [Google Scholar] [CrossRef]
- Mascot Search Engine|Protein Identification Software for Mass Spec Data, (n.d.). Available online: http://www.matrixscience.com/ (accessed on 27 June 2022).
- Amanat, F.; White, K.M.; Miorin, L.; Strohmeier, S.; McMahon, M.; Meade, P.; Liu, W.C.; Albrecht, R.A.; Simon, V.; Martinez-Sobrido, L.; et al. An In Vitro Microneutralization Assay for SARS-CoV-2 Serology and Drug Screening. Curr. Protoc. Microbiol. 2020, 58, e108. [Google Scholar] [CrossRef]
- Kozlovskaya, L.; Piniaeva, A.; Ignatyev, G.; Selivanov, A.; Shishova, A.; Kovpak, A.; Gordeychuk, I.; Ivin, Y.; Berestovskaya, A.; Prokhortchouk, E.; et al. Isolation and phylogenetic analysis of SARS-CoV-2 variants collected in Russia during the COVID-19 outbreak. Int. J. Infect. Dis. 2020, 99, 40–46. [Google Scholar] [CrossRef]
- Ksenafontov, A.D.; Pisareva, M.M.; Eder, V.A.; Musaeva, T.D.; Timofeeva, M.M.; Kiseleva, I.V. Comparing of level of circulation of respiratory viruses during epidemic seasons 2015–2020 and COVID-19 pandemic in Saint Petersburg. Med. Acad. J. 2021, 21, 113–117. [Google Scholar] [CrossRef]
- Szymczak, A.; Jedruchniewicz, N.; Torelli, A.; Kaczmarzyk-Radka, A.; Coluccio, R.; Klak, M.; Konieczny, A.; Ferenc, S.; Witkiewicz, W.; Montomoli, E.; et al. Antibodies specific to SARS-CoV-2 proteins N, S and E in COVID-19 patients in the normal population and in historical samples. J. Gen. Virol. 2021, 102, 001692. [Google Scholar] [CrossRef]
- Turkkan, A.; Saglik, I.; Turan, C.; Sahin, A.; Akalin, H.; Ener, B.; Kara, A.; Celebi, S.; Sahin, E.; Hacimustafaoglu, M. Nine-month course of SARS-CoV-2 antibodies in individuals with COVID-19 infection. Ir. J. Med. Sci. 2022. [Google Scholar] [CrossRef]
- Kudryavtsev, I.; Rubinstein, A.; Golovkin, A.; Kalinina, O.; Vasilyev, K.; Rudenko, L.; Isakova-Sivak, I. Dysregulated Immune Responses in SARS-CoV-2-Infected Patients: A Comprehensive Overview. Viruses 2022, 14, 1082. [Google Scholar] [CrossRef]
- Tutukina, M.; Kaznadzey, A.; Kireeva, M.; Mazo, I. IgG Antibodies Develop to Spike but Not to the Nucleocapsid Viral Protein in Many Asymptomatic and Light COVID-19 Cases. Viruses 2021, 13, 1945. [Google Scholar] [CrossRef]
- Brochot, E.; Demey, B.; Touze, A.; Belouzard, S.; Dubuisson, J.; Schmit, J.L.; Duverlie, G.; Francois, C.; Castelain, S.; Helle, F. Anti-spike, Anti-nucleocapsid and Neutralizing Antibodies in SARS-CoV-2 Inpatients and Asymptomatic Individuals. Front. Microbiol. 2020, 11, 584251. [Google Scholar] [CrossRef]
- Zhao, J.; Yuan, Q.; Wang, H.; Liu, W.; Liao, X.; Su, Y.; Wang, X.; Yuan, J.; Li, T.; Li, J.; et al. Antibody Responses to SARS-CoV-2 in Patients With Novel Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 2027–2034. [Google Scholar] [CrossRef]
- Uysal, E.B.; Gumus, S.; Bektore, B.; Bozkurt, H.; Gozalan, A. Evaluation of antibody response after COVID-19 vaccination of healthcare workers. J. Med. Virol. 2022, 94, 1060–1066. [Google Scholar] [CrossRef]
- Wei, J.; Pouwels, K.B.; Stoesser, N.; Matthews, P.C.; Diamond, I.; Studley, R.; Rourke, E.; Cook, D.; Bell, J.I.; Newton, J.N.; et al. Antibody responses and correlates of protection in the general population after two doses of the ChAdOx1 or BNT162b2 vaccines. Nat. Med. 2022, 28, 1072–1082. [Google Scholar] [CrossRef]
- Heyming, T.W.; Nugent, D.; Tongol, A.; Knudsen-Robbins, C.; Hoang, J.; Schomberg, J.; Bacon, K.; Lara, B.; Sanger, T. Rapid antibody testing for SARS-CoV-2 vaccine response in pediatric healthcare workers. Int. J. Infect. Dis. 2021, 113, 1–6. [Google Scholar] [CrossRef]
- Ayón-Núñez, D.A.; Cervantes-Torres, J.; Cabello-Gutiérrez, C.; Rosales-Mendoza, S.; Rios-Valencia, D.; Huerta, L.; Bobes, R.J.; Carrero, J.C.; Segura-Velázquez, R.; Fierro, N.A.; et al. An RBD-Based Diagnostic Method Useful for the Surveillance of Protective Immunity against SARS-CoV-2 in the Population. Diagnostics 2022, 12, 1629. [Google Scholar] [CrossRef]
- Peterhoff, D.; Glück, V.; Vogel, M.; Schuster, P.; Schütz, A.; Neubert, P.; Albert, V.; Frisch, S.; Kiessling, M.; Pervan, P.; et al. A highly specific and sensitive serological assay detects SARS-CoV-2 antibody levels in COVID-19 patients that correlate with neutralization. Infection 2021, 49, 75–82. [Google Scholar] [CrossRef]
- Mehdi, F.; Chattopadhyay, S.; Thiruvengadam, R.; Yadav, S.; Kumar, M.; Sinha, S.K.; Goswami, S.; Kshetrapal, P.; Wadhwa, N.; Chandramouli Natchu, U.; et al. Development of a Fast SARS-CoV-2 IgG ELISA, Based on Receptor-Binding Domain, and Its Comparative Evaluation Using Temporally Segregated Samples From RT-PCR Positive Individuals. Front. Microbiol. 2020, 11, 618097. [Google Scholar] [CrossRef]
- Wagner, A.; Guzek, A.; Ruff, J.; Jasinska, J.; Scheikl, U.; Zwazl, I.; Kundi, M.; Stockinger, H.; Farcet, M.R.; Kreil, T.R.; et al. Neutralising SARS-CoV-2 RBD-specific antibodies persist for at least six months independently of symptoms in adults. Commun. Med. 2021, 1, 13. [Google Scholar] [CrossRef]
- Greaney, A.J.; Starr, T.N.; Barnes, C.O.; Weisblum, Y.; Schmidt, F.; Caskey, M.; Gaebler, C.; Cho, A.; Agudelo, M.; Finkin, S.; et al. Mapping mutations to the SARS-CoV-2 RBD that escape binding by different classes of antibodies. Nat. Commun. 2021, 12, 4196. [Google Scholar] [CrossRef]
- Bozdaganyan, M.E.; Shaitan, K.V.; Kirpichnikov, M.P.; Sokolova, O.S.; Orekhov, P.S. Computational Analysis of Mutations in the Receptor-Binding Domain of SARS-CoV-2 Spike and Their Effects on Antibody Binding. Viruses 2022, 14, 295. [Google Scholar] [CrossRef]
- Ding, C.; He, J.; Zhang, X.; Jiang, C.; Sun, Y.; Zhang, Y.; Chen, Q.; He, H.; Li, W.; Xie, J.; et al. Crucial Mutations of Spike Protein on SARS-CoV-2 Evolved to Variant Strains Escaping Neutralization of Convalescent Plasmas and RBD-Specific Monoclonal Antibodies. Front. Immunol. 2021, 12, 693775. [Google Scholar] [CrossRef]
- Koerber, N.; Priller, A.; Yazici, S.; Bauer, T.; Cheng, C.C.; Mijocevic, H.; Wintersteller, H.; Jeske, S.; Vogel, E.; Feuerherd, M.; et al. Dynamics of spike-and nucleocapsid specific immunity during long-term follow-up and vaccination of SARS-CoV-2 convalescents. Nat. Commun. 2022, 13, 153. [Google Scholar] [CrossRef]
- Gonzalez Lopez Ledesma, M.M.; Sanchez, L.; Ojeda, D.S.; Oviedo Rouco, S.; Rossi, A.H.; Varese, A.; Mazzitelli, I.; Pascuale, C.A.; Miglietta, E.A.; Rodriguez, P.E.; et al. Longitudinal Study after Sputnik V Vaccination Shows Durable SARS-CoV-2 Neutralizing Antibodies and Reduced Viral Variant Escape to Neutralization over Time. mBio 2022, 13, e03442-21. [Google Scholar] [CrossRef]
- Chahla, R.E.; Tomas-Grau, R.H.; Cazorla, S.I.; Ploper, D.; Vera Pingitore, E.; Lopez, M.A.; Aznar, P.; Alcorta, M.E.; Velez, E.; Stagnetto, A.; et al. Long-term analysis of antibodies elicited by SPUTNIK V: A prospective cohort study in Tucuman, Argentina. Lancet Reg. Health Am. 2022, 6, 100123. [Google Scholar] [CrossRef]
- Blanco, S.; Salome Konigheim, B.; Diaz, A.; Spinsanti, L.; Javier Aguilar, J.; Elisa Rivarola, M.; Beranek, M.; Collino, C.; MinSalCba Working, G.; Fcm-Unc Working, G.; et al. Evaluation of the Gam-COVID-Vac and vaccine-induced neutralizing response against SARS-CoV-2 lineage P.1 variant in an Argentinean cohort. Vaccine 2022, 40, 811–818. [Google Scholar] [CrossRef]
- Dutta, N.K.; Mazumdar, K.; Gordy, J.T. The Nucleocapsid Protein of SARS-CoV-2: A Target for Vaccine Development. J. Virol. 2020, 94, e00647-20. [Google Scholar] [CrossRef]
- Thakur, S.; Sasi, S.; Pillai, S.G.; Nag, A.; Shukla, D.; Singhal, R.; Phalke, S.; Velu, G.S.K. SARS-CoV-2 Mutations and Their Impact on Diagnostics, Therapeutics and Vaccines. Front. Med. 2022, 9, 815389. [Google Scholar] [CrossRef]
- Paul, G.; Strnad, P.; Wienand, O.; Krause, U.; Plecko, T.; Effenberger-Klein, A.; Giel, K.E.; Junne, F.; Galante-Gottschalk, A.; Ehehalt, S.; et al. The humoral immune response more than one year after SARS-CoV-2 infection: Low detection rate of anti-nucleocapsid antibodies via Euroimmun ELISA. Infection 2022. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rak, A.; Donina, S.; Zabrodskaya, Y.; Rudenko, L.; Isakova-Sivak, I. Cross-Reactivity of SARS-CoV-2 Nucleocapsid-Binding Antibodies and Its Implication for COVID-19 Serology Tests. Viruses 2022, 14, 2041. https://doi.org/10.3390/v14092041
Rak A, Donina S, Zabrodskaya Y, Rudenko L, Isakova-Sivak I. Cross-Reactivity of SARS-CoV-2 Nucleocapsid-Binding Antibodies and Its Implication for COVID-19 Serology Tests. Viruses. 2022; 14(9):2041. https://doi.org/10.3390/v14092041
Chicago/Turabian StyleRak, Alexandra, Svetlana Donina, Yana Zabrodskaya, Larisa Rudenko, and Irina Isakova-Sivak. 2022. "Cross-Reactivity of SARS-CoV-2 Nucleocapsid-Binding Antibodies and Its Implication for COVID-19 Serology Tests" Viruses 14, no. 9: 2041. https://doi.org/10.3390/v14092041





