A Conserved Stem-Loop Structure within ORF5 Is a Frequent Recombination Hotspot for Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) with a Particular Modified Live Virus (MLV) Strain
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells
2.2. Virus Isolation by Cell Culture
2.3. RT-PCR
2.4. Virus Propagation and RNA Extraction
2.5. cDNA Preparation and Sequencing
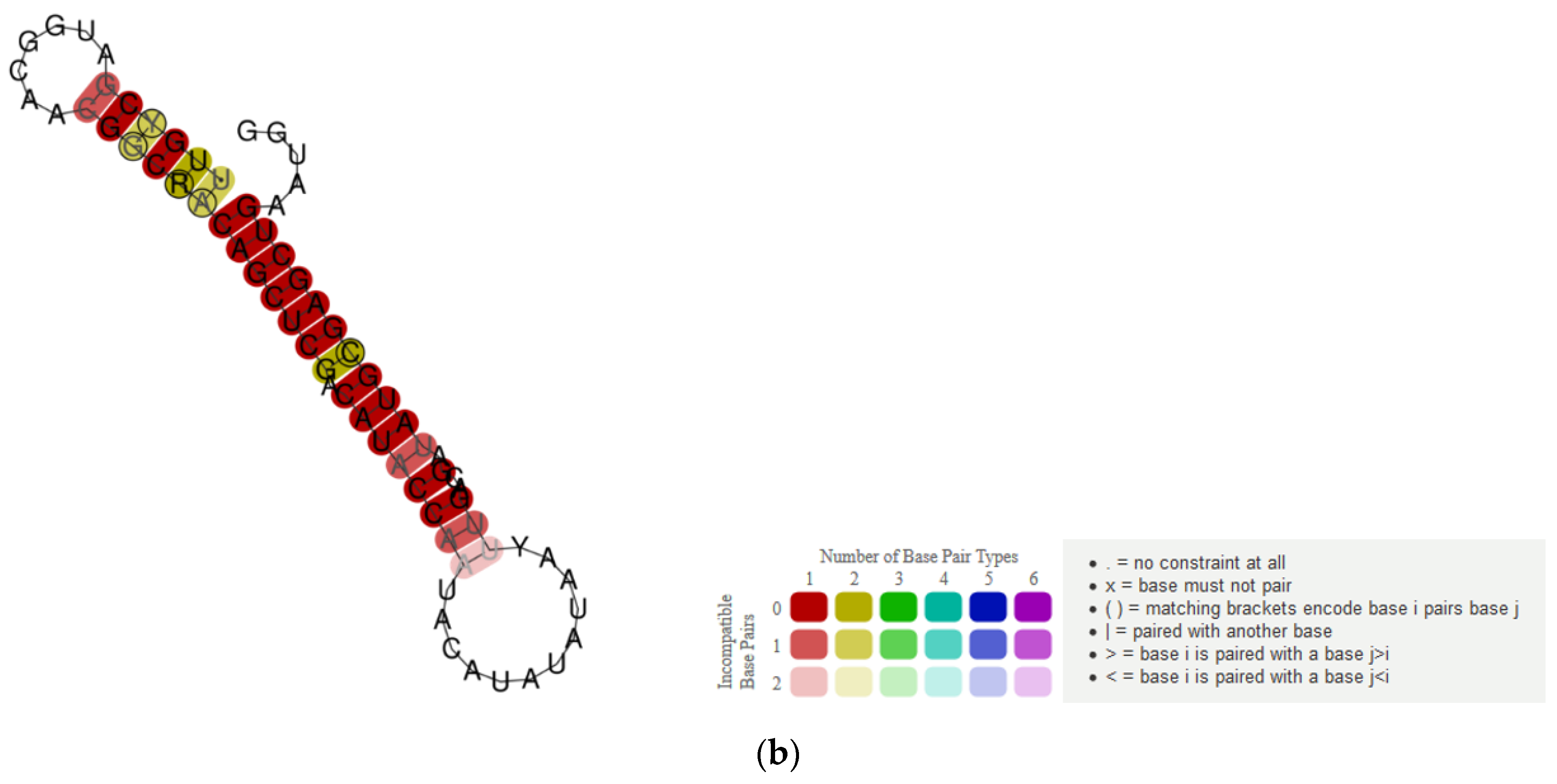
2.6. Recombination Analysis and RNA Structure Predictions
3. Results
3.1. Isolation and Sequencing of Chimeric PRRSV Strains
3.2. Anamnesis and Clinical Findings in the Affected Farms
3.3. PRRSV Isolates Are Recombinant Viruses Harboring ORF5–3′ Sequences from a Particular Modified Life Vaccine Strain
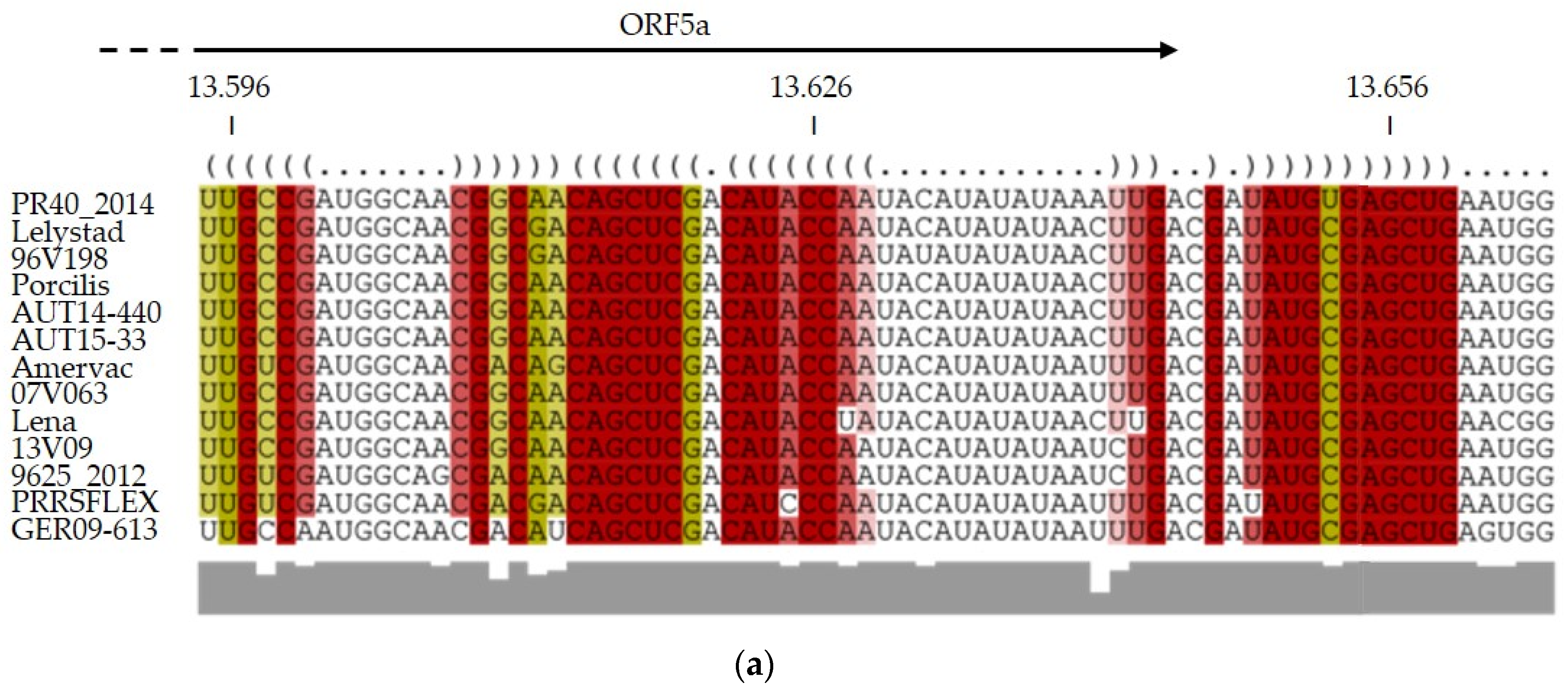
3.4. RNA Structure Predictions Reveal a Conserved Stem Loop Upstream of ORF5a
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Primer | Sequence | Forward/Reverse | Binding at nt |
|---|---|---|---|
| PRS328 | GYGAWCTYCAAGTTTAYGAGC | forward | 527–547 |
| PRS329 | GAGCTCCAARCAGGCCATG | forward | 2040–2059 |
| PRS330 | CCRATACTCGGATGCCTTCC | forward | 3506–3527 |
| PRS331 | CCACGTGGTGCRGCTGCTG | forward | 4993–5011 |
| PRS332 | AYGTRATYGTTCTGCTTGGGC | forward | 6533–6553 |
| PRS333 | TGCCTGGGGTCCTACGCCT | forward | 7984–8002 |
| PRS334 | GAYGATGATGTYATYTACACACC | forward | 9636–9658 |
| PRS335 | CTCTCACCGATGTGTACCTY | forward | 11,002–11,021 |
| PRS336 | CGTCCGGGTACGAYAAYCTY | forward | 12,502–12,521 |
| PRS337 | GTGTCWCGCGGCCGACTCYTG | forward | 14,004–14,024 |
| PRS338 | TGGTCRGACACGTGCATGGAG | reverse | 606–626 |
| PRS339 | RGGCCTTKGAGGAKGGRAG | reverse | 2081–2098 |
| PRS340 | GCCATCCAAGAACCAAAAACAC | reverse | 3572–3593 |
| PRS341 | CTGAARGCACCTTCRAGRAGGG | reverse | 5105–5126 |
| PRS342 | CATTRATRTCGAGGATGGATCC | reverse | 6565–6586 |
| PRS343 | TGGCCATTRAYCCCTGCCA | reverse | 8083–8102 |
| PRS344 | GGAATACCTRCAAACTTTRAGAGC | reverse | 9690–9713 |
| PRS345 | TTCCAGCATTTTGAYGCCGTC | reverse | 11,051–11,071 |
| PRS346 | MGGATGGAAYTGGGCCGCT | reverse | 12,569–12,588 |
| PRS347 | AAATGCACATATGTCATGTAYCC | reverse | 14,070–14,092 |
References
- Nelsen, C.J.; Murtaugh, M.P.; Faaberg, K.S. Porcine Reproductive and Respiratory Syndrome Virus Comparison: Divergent Evolution on Two Continents. J. Virol. 1999, 73, 270–280. [Google Scholar] [CrossRef] [Green Version]
- Chang, C.-C.; Yoon, K.-J.; Zimmerman, J.J.; Harmon, K.M.; Dixon, P.M.; Dvorak, C.M.T.; Murtaugh, M.P. Evolution of Porcine Reproductive and Respiratory Syndrome Virus during Sequential Passages in Pigs. J. Virol. 2002, 76, 4750–4763. [Google Scholar] [CrossRef] [Green Version]
- Lauber, C.; Goeman, J.J.; de Parquet, M.C.; Thi Nga, P.; Snijder, E.J.; Morita, K.; Gorbalenya, A.E. The Footprint of Genome Architecture in the Largest Genome Expansion in RNA Viruses. PLoS Pathog. 2013, 9, e1003500. [Google Scholar] [CrossRef] [Green Version]
- Goldberg, T.L.; Lowe, J.F.; Milburn, S.M.; Firkins, L.D. Quasispecies Variation of Porcine Reproductive and Respiratory Syndrome Virus during Natural Infection. Virology 2003, 317, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Conzelmann, K.K.; Visser, N.; Van Woensel, P.; Thiel, H.J. Molecular Characterization of Porcine Reproductive and Respiratory Syndrome Virus, a Member of the Arterivirus Group. Virology 1993, 193, 329–339. [Google Scholar] [CrossRef] [Green Version]
- Lunney, J.K.; Fang, Y.; Ladinig, A.; Chen, N.; Li, Y.; Rowland, B.; Renukaradhya, G.J. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV): Pathogenesis and Interaction with the Immune System. Annu. Rev. Anim. Biosci. 2016, 4, 129–154. [Google Scholar] [CrossRef]
- Neumann, E.J.; Kliebenstein, J.B.; Johnson, C.D.; Mabry, J.W.; Bush, E.J.; Seitzinger, A.H.; Green, A.L.; Zimmerman, J.J. Assessment of the Economic Impact of Porcine Reproductive and Respiratory Syndrome on Swine Production in the United States. J. Am. Vet. Med. Assoc. 2005, 227, 385–392. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, H.S.; Oleksiewicz, M.B.; Forsberg, R.; Stadejek, T.; Bøtner, A. Reversion of a Live Porcine Reproductive and Respiratory Syndrome Virus Vaccine Investigated by Parallel Mutations. J. Gen. Virol. 2001, 82, 1263–1272. [Google Scholar] [CrossRef]
- Li, B.; Fang, L.; Xu, Z.; Liu, S.; Gao, J.; Jiang, Y.; Chen, H.; Xiao, S. Recombination in Vaccine and Circulating Strains of Porcine Reproductive and Respiratory Syndrome Viruses. Emerg. Infect. Dis. 2009, 15, 2032–2035. [Google Scholar] [CrossRef]
- Marton, S.; Szalay, D.; Kecskeméti, S.; Forró, B.; Olasz, F.; Zádori, Z.; Szabó, I.; Molnár, T.; Bányai, K. Bálint Coding-Complete Sequence of a Vaccine-Derived Recombinant Porcine Reproductive and Respiratory Syndrome Virus Strain Isolated in Hungary. Arch. Virol. 2019, 164, 2605–2608. [Google Scholar] [CrossRef]
- Vandenbussche, F.; Mathijs, E.; Tignon, M.; Vandersmissen, T.; Cay, A.B. WGS- versus ORF5-Based Typing of PRRSV: A Belgian Case Study. Viruses 2021, 13, 2419. [Google Scholar] [CrossRef]
- Pérez-Losada, M.; Arenas, M.; Galán, J.C.; Palero, F.; González-Candelas, F. Recombination in Viruses: Mechanisms, Methods of Study, and Evolutionary Consequences. Infect. Genet. Evol. 2015, 30, 296–307. [Google Scholar] [CrossRef] [Green Version]
- Rabadan, R.; Levine, A.J.; Krasnitz, M. Non-Random Reassortment in Human Influenza A Viruses. Influ. Other Respir. Viruses 2008, 2, 9–22. [Google Scholar] [CrossRef] [Green Version]
- Hwang, C.K.; Svarovskaia, E.S.; Pathak, V.K. Dynamic Copy Choice: Steady State between Murine Leukemia Virus Polymerase and Polymerase-Dependent RNase H Activity Determines Frequency of in Vivo Template Switching. Proc. Natl. Acad. Sci. USA 2001, 98, 12209–12214. [Google Scholar] [CrossRef] [Green Version]
- Malim, M.H.; Emerman, M. HIV-1 Sequence Variation: Drift, Shift, and Attenuation. Cell 2001, 104, 469–472. [Google Scholar] [CrossRef] [Green Version]
- Khatchikian, D.; Orlich, M.; Rott, R. Increased Viral Pathogenicity after Insertion of a 28S Ribosomal RNA Sequence into the Haemagglutinin Gene of an Influenza Virus. Nature 1989, 340, 156–157. [Google Scholar] [CrossRef]
- Liu, Y.; Li, J.; Yang, J.; Zeng, H.; Guo, L.; Ren, S.; Sun, W.; Chen, Z.; Cong, X.; Shi, J.; et al. Emergence of Different Recombinant Porcine Reproductive and Respiratory Syndrome Viruses, China. Sci. Rep. 2018, 8, 4118. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Lin, S.; Quan, D.; Wang, H.; Huang, J.; Wang, Y.; Ren, T.; Ouyang, K.; Chen, Y.; Huang, W.; et al. Full Genomic Analysis of New Variants of Porcine Reproductive and Respiratory Syndrome Virus Revealed Multiple Recombination Events Between Different Lineages and Sublineages. Front. Vet. Sci. 2020, 7, 603. [Google Scholar] [CrossRef]
- Kvisgaard, L.K.; Kristensen, C.S.; Ryt-Hansen, P.; Pedersen, K.; Stadejek, T.; Trebbien, R.; Andresen, L.O.; Larsen, L.E. A Recombination between Two Type 1 Porcine Reproductive and Respiratory Syndrome Virus (PRRSV-1) Vaccine Strains Has Caused Severe Outbreaks in Danish Pigs. Transbound. Emerg. Dis. 2020, 67, 1786–1796. [Google Scholar] [CrossRef] [Green Version]
- Kreutzmann, H.; Stadler, J.; Knecht, C.; Sassu, E.K.; Ruczizka, U.; Zablotski, Y. Phenotypic Characterization of a Virulent PRRSV-1 Isolate in a Reproductive Model With and Without Prior Heterologous Modified Live PRRSV-1 Vaccination. Front. Vet. Sci. 2022, 9, 820233. [Google Scholar] [CrossRef]
- Lole, K.S.; Bollinger, R.C.; Paranjape, R.S.; Gadkari, D.; Kulkarni, S.S.; Novak, N.G.; Ingersoll, R.; Sheppard, H.W.; Ray, S.C. Full-Length Human Immunodeficiency Virus Type 1 Genomes from Subtype C-Infected Seroconverters in India, with Evidence of Intersubtype Recombination. J. Virol. 1999, 73, 152–160. [Google Scholar] [CrossRef] [Green Version]
- Martin, D.P.; Varsani, A.; Roumagnac, P.; Botha, G.; Maslamoney, S.; Schwab, T. RDP5: A Computer Program for Analyzing Recombination in, and Removing Signals of Recombination from, Nucleotide Sequence Datasets. Virus Evol. 2020, 7, veaa087. [Google Scholar] [CrossRef] [PubMed]
- Will, S.; Joshi, T.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. LocARNA-P: Accurate Boundary Prediction and Improved Detection of Structural RNAs. RNA 2012, 18, 900–914. [Google Scholar] [CrossRef] [Green Version]
- Raden, M.; Ali, S.M.; Alkhnbashi, O.S.; Busch, A.; Costa, F.; Davis, J.A. Freiburg RNA Tools: A Central Online Resource for RNA-Focused Research and Teaching. Nucleic Acids Res. 2018, 46, W25–W29. [Google Scholar] [CrossRef] [PubMed]
- Will, S.; Reiche, K.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. Inferring Non-Coding RNA Families and Classes by Means of Genome-Scale Structure-Based Clustering. PLoS Comput. Biol. 2007, 3, e65. [Google Scholar] [CrossRef] [PubMed]
- Sinn, L.J.; Zieglowski, L.; Koinig, H.; Lamp, B.; Jansko, B.; Mößlacher, G.; Riedel, C.; Hennig-Pauka, I.; Rümenapf, T. Characterization of Two Austrian Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Field Isolates Reveals Relationship to East Asian Strains. Vet. Res. 2016, 47, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, S.W.C.; Lynch, E.C.; Leason, K.R.; Court, D.L.; Shapiro, B.A.; Friedman, D.I. Functional Importance of Sequence in the Stem-Loop of a Transcription Terminator. Science 1991, 254, 1205–1207. [Google Scholar] [CrossRef] [PubMed]
- Wenhui, L.; Zhongyan, W.; Guanqun, Z.; Zhili, L.; JingYun, M.; Qingmei, X. Complete Genome Sequence of a Novel Variant Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) Strain: Evidence for Recombination between Vaccine and Wild-Type PRRSV Strains. J. Virol. 2012, 254, 1205–1207. [Google Scholar] [CrossRef] [Green Version]
- Kristensen, C.S.; Christiansen, M.G.; Pedersen, K.; Larsen, L.E. Production Losses Five Months after Outbreak with a Recombinant of Two PRRSV Vaccine Strains in 13 Danish Sow Herds. Porc. Health Manag. 2020, 6, 26. [Google Scholar] [CrossRef]
- Zhao, D.; Yang, B.; Yuan, X.; Shen, C.; Zhang, D.; Shi, X.; Zhang, T.; Cui, H.; Yang, J.; Chen, X.; et al. Advanced Research in Porcine Reproductive and Respiratory Syndrome Virus Co-Infection With Other Pathogens in Swine. Front. Vet. Sci. 2021, 8, 982. [Google Scholar] [CrossRef]
- Sagripanti, J.L.; Zandomeni, R.O.; Weinmann, R. The Cap Structure of Simian Hemorrhagic Fever Virion RNA. Virology 1986, 151, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Gao, F.; Wei, Z.; Liu, P.; Liu, C.; Zheng, H.; Li, Y.; Lin, T.; Yuan, S. A 5′-Proximal Stem-Loop Structure of 5′ Untranslated Region of Porcine Reproductive and Respiratory Syndrome Virus Genome Is Key for Virus Replication. Virol. J. 2011, 8, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, C.L.; Lai, M.M. Requirement of the 5′-End Genomic Sequence as an Upstream Cis-Acting Element for Coronavirus Subgenomic MRNA Transcription. J. Virol. 1994, 68, 4727–4737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.-J.; Yun, S.-I.; Kang, S.-Y.; Lee, Y.-M. Identification of 5′ and 3′ Cis-Acting Elements of the Porcine Reproductive and Respiratory Syndrome Virus: Acquisition of Novel 5′ AU-Rich Sequences Restored Replication of a 5′-Proximal 7-Nucleotide Deletion Mutant. J. Virol. 2006, 80, 723–736. [Google Scholar] [CrossRef] [Green Version]
- Chang, R.Y.; Krishnan, R.; Brian, D.A. The UCUAAAC Promoter Motif Is Not Required for High-Frequency Leader Recombination in Bovine Coronavirus Defective Interfering RNA. J. Virol. 1996, 70, 2720–2729. [Google Scholar] [CrossRef] [Green Version]
- Van Marle, G.; Dobbe, J.C.; Gultyaev, A.P.; Luytjes, W.; Spaan, W.J.M.; Snijder, E.J. Arterivirus Discontinuous MRNA Transcription Is Guided by Base Pairing between Sense and Antisense Transcription-Regulating Sequences. Proc. Natl. Acad. Sci. USA 1999, 96, 12056–12061. [Google Scholar] [CrossRef] [Green Version]
- Sinn, L.J.; Klingler, E.; Lamp, B.; Brunthaler, R.; Weissenböck, H.; Rümenapf, T. Emergence of a virulent porcine reproductive and respiratory syndrome virus (PRRSV) 1 strain in Lower Austria. Porc. Health Manag. 2016, 2, 28. [Google Scholar] [CrossRef] [Green Version]
- Duerlinger, S.; Knecht, C.; Sawyer, S.; Balka, G.; Zaruba, M.; Ruemenapf, T.; Kraft, C.; Rathkjen, P.H.; Ladinig, A. Efficacy of a Modified Live Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) Vaccine against Experimental Infection with PRRSV AUT15-33 in Weaned Piglets. Vaccines 2022, 10, 934. [Google Scholar] [CrossRef]
- Holtkamp, D.J.; Yeske, P.E.; Polson, D.D.; Melody, J.L.; Philips, R.C. A Prospective Study Evaluating Duration of Swine Breeding Herd PRRS Virus-Free Status and Its Relationship with Measured Risk. Prev. Vet. Med. 2010, 96, 186–193. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mötz, M.; Stadler, J.; Kreutzmann, H.; Ladinig, A.; Lamp, B.; Auer, A.; Riedel, C.; Rümenapf, T. A Conserved Stem-Loop Structure within ORF5 Is a Frequent Recombination Hotspot for Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) with a Particular Modified Live Virus (MLV) Strain. Viruses 2023, 15, 258. https://doi.org/10.3390/v15010258
Mötz M, Stadler J, Kreutzmann H, Ladinig A, Lamp B, Auer A, Riedel C, Rümenapf T. A Conserved Stem-Loop Structure within ORF5 Is a Frequent Recombination Hotspot for Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) with a Particular Modified Live Virus (MLV) Strain. Viruses. 2023; 15(1):258. https://doi.org/10.3390/v15010258
Chicago/Turabian StyleMötz, Marlene, Julia Stadler, Heinrich Kreutzmann, Andrea Ladinig, Benjamin Lamp, Angelika Auer, Christiane Riedel, and Till Rümenapf. 2023. "A Conserved Stem-Loop Structure within ORF5 Is a Frequent Recombination Hotspot for Porcine Reproductive and Respiratory Syndrome Virus 1 (PRRSV-1) with a Particular Modified Live Virus (MLV) Strain" Viruses 15, no. 1: 258. https://doi.org/10.3390/v15010258






