Recent Advances in Structural Studies of Single-Stranded RNA Bacteriophages
Abstract
:1. Introduction
| Phages | Genus | Hosts | Receptors |
|---|---|---|---|
| MS2 | Emesvirus | E. coli | Conjugative F pili |
| Qβ | Qubevirus | E. coli | Conjugative F pili |
| PP7 | Pepevirus | P. aeruginosa | Type IV pili |
| LeviOr01 | Pepevirus | P. aeruginosa | Type IV pili |
| AP205 | Apeevirus | Acinetobacter spp. | Type IV pili |
| ϕCb5 | Cebevirus | C. crescentus | Type IV Tad pili |
| PRR1 | Perrunavirus | Pseudomonas, Salmonella, Vibrio, Escherichia | Conjugative P pili |
| M | Empivirus | Escherichia, Salmonella, Klebsiella, Proteus and Serratia | Conjugative M pili |
| C-1 | Cunavirus | Escherichia, Salmonella, Proteus and Serratia | Conjugative C pili |
| Hgal1 | Hagavirus | Escherichia, Citrobacter, Klebsiella, Enterobacter | Conjugative H pili |
2. The Known Structures of ssRNA Phage Capsids: VLPs and Mature Virions
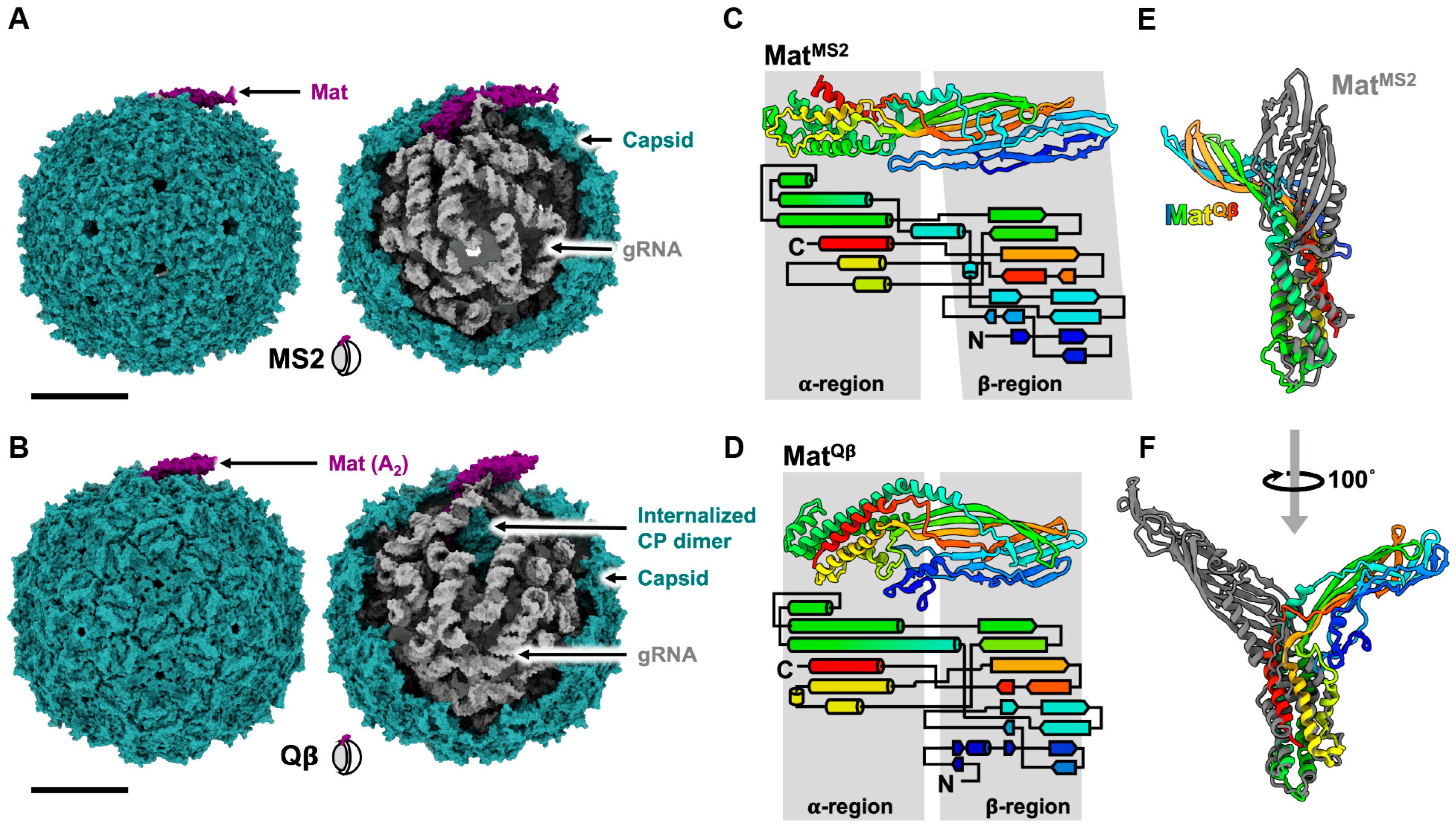
3. The Interactions between Host Receptors and ssRNA Phages
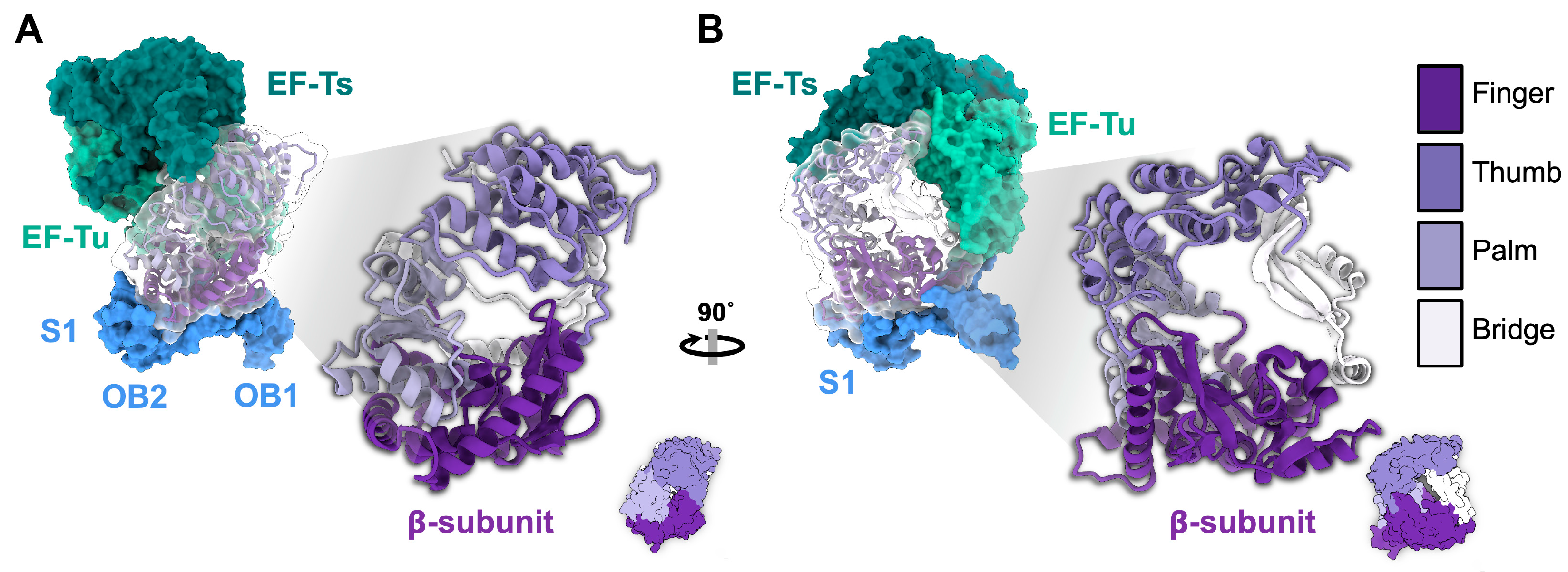
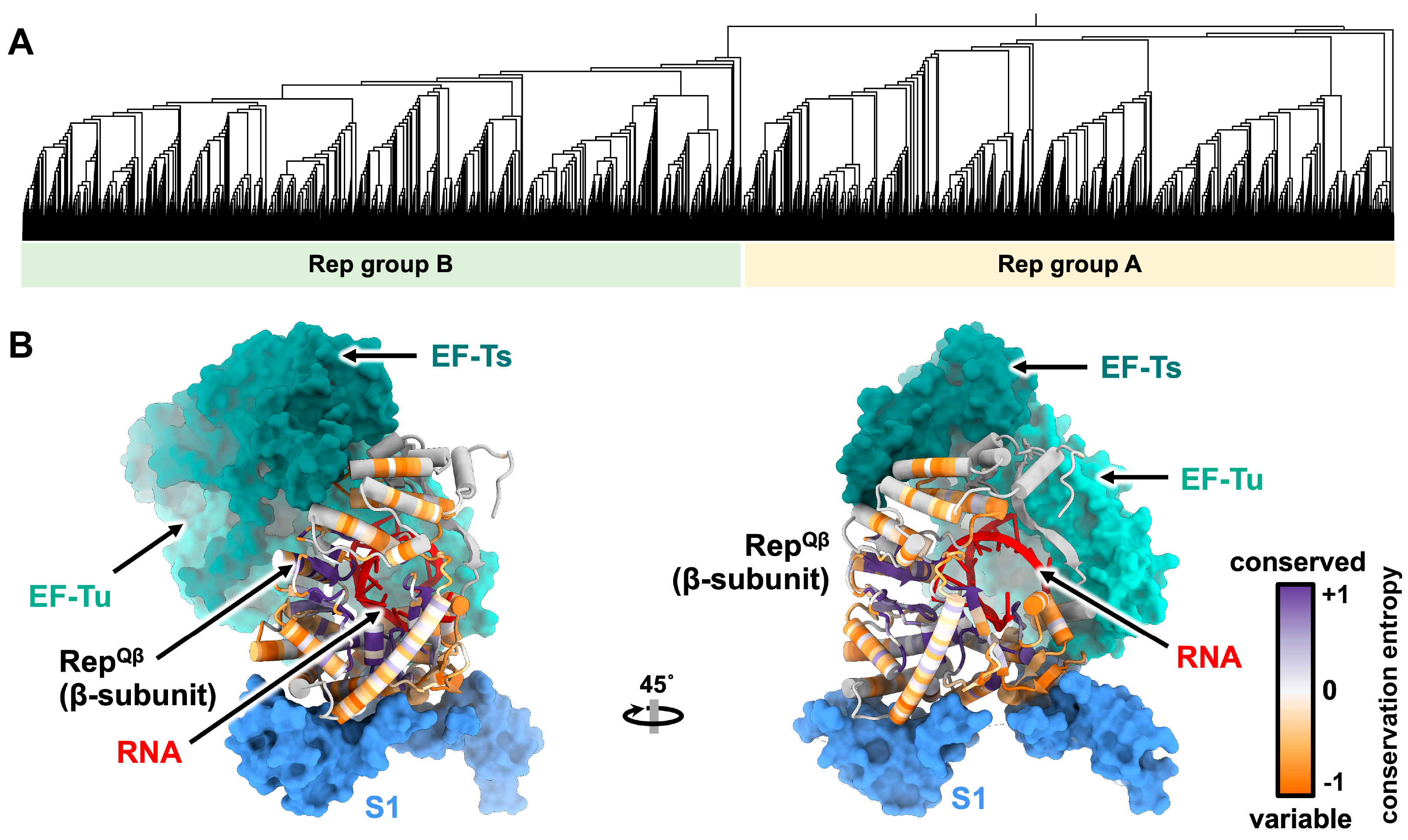
4. The Genome Replication of ssRNA Phages
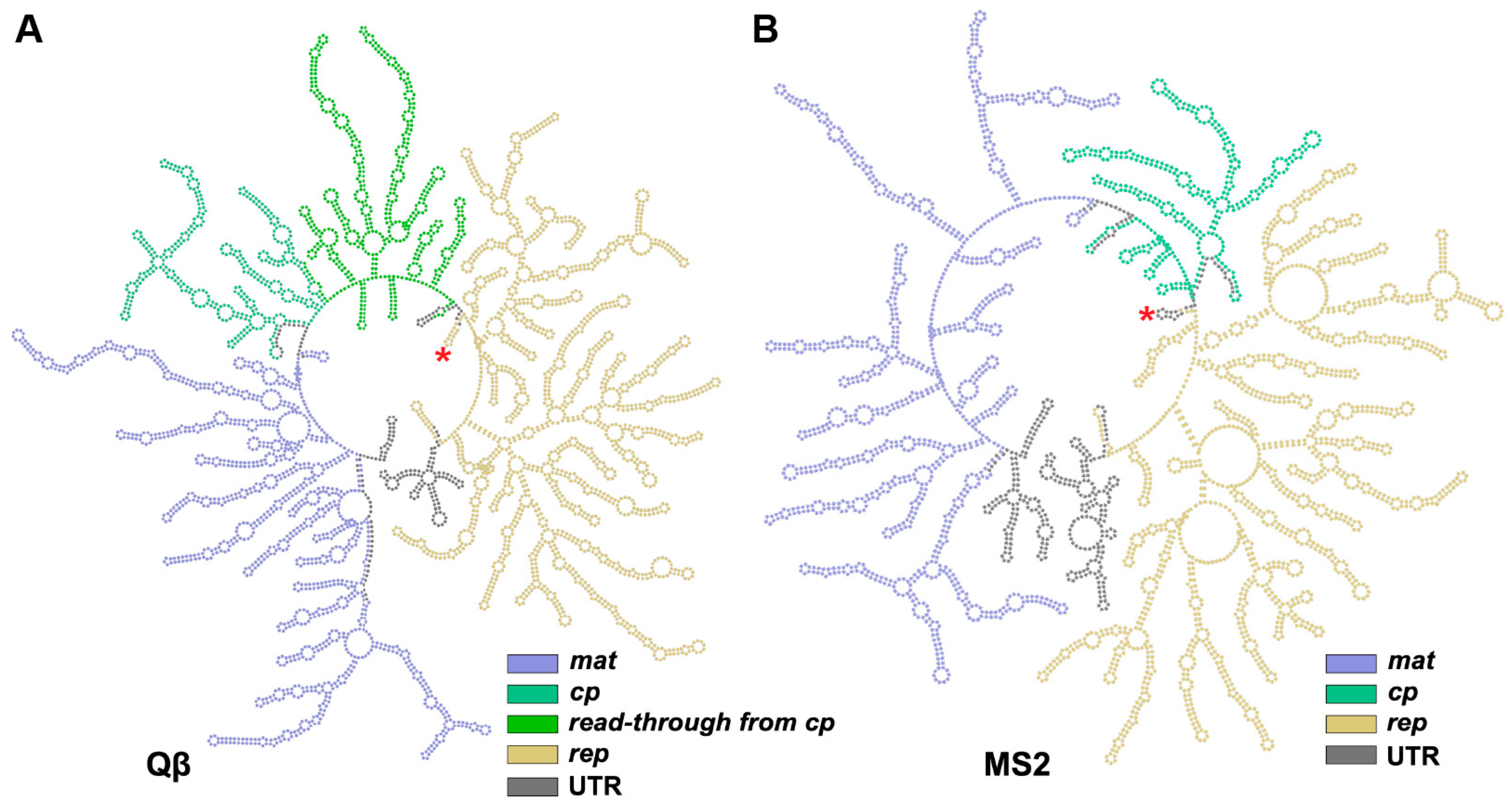
5. The Genome Packaging and Viral Assembly of ssRNA Phages
6. The Lysis of the Host by ssRNA Phages
7. Conclusions and Future Directions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Phages | PDB# | Method | Brief Description |
|---|---|---|---|
| Culturable ssRNA phages | |||
| MS2 | 2MS2 | X-ray | T = 3 VLP |
| 1BMS | X-ray | T = 3 VLP (P78N) | |
| 1MST | X-ray | T = 3 VLP (E76D) | |
| 1ZDI | X-ray | CP/RNA complex, WT RNA | |
| 1ZDH | X-ray | CP/RNA complex, C-5 variant RNA (U→C mutant) | |
| 1AQ3 | X-ray | CP(T59S)/RNA complex | |
| 1AQ4 | X-ray | CP(T45A)/RNA complex | |
| 1MVA | X-ray | T = 3 VLP, CP(T45A) | |
| 1MVB | X-ray | T = 3 VLP, CP(T59S) | |
| 1ZDJ | X-ray | CP/RNA complex, loop RNA (5′-GGAUCACC-3′), shorter RNA operator derived from WT RNA operator | |
| 1ZDK | X-ray | CP/RNA complex, clamp RNA, elongated stem of C-5 variant RNA operator | |
| 6MSF | X-ray | CP/RNA complex, F6 RNA, derived from WT and C-5 version | |
| 5MSF | X-ray | CP/RNA complex, F5 RNA, derived from WT and C-5 version | |
| 7MSF | X-ray | CP/RNA complex, F7 RNA, derived from WT and C-5 version | |
| 1U1Y | X-ray | CP/RNA complex, F5 RNA with 2-aminopurine at -10 position | |
| 2BU1 | X-ray | CP/RNA complex, RNA operator with 5-bromouracil at -5 position | |
| 2C4Q | X-ray | CP/RNA complex, RNA operator with pyridin-2-one at -5 position | |
| 2C4Y | X-ray | CP/RNA complex, RNA operator with 2-thiouracil at -5 position | |
| 2C4Z | X-ray | CP/RNA complex, RNA operator with 2-thiouracil at -5 and -6 position | |
| 2C50 | X-ray | CP/RNA complex, RNA operator with adenine at -5 position | |
| 2C51 | X-ray | CP/RNA complex, RNA operator with guanine at -5 position | |
| 2BNY | X-ray | CP(N87A)/ MS2 RNA complex | |
| 2BQ5 | X-ray | CP(N87A/E89K)/MS2 RNA complex | |
| 2BS0 | X-ray | CP(N87A/E89K)/variant Qβ RNA complex | |
| 2BS1 | X-ray | CP(N87A/E89K)/Qβ RNA complex | |
| 1ZSE | X-ray | CP(N87S)/Qβ RNA complex | |
| 2B2D | X-ray | CP(N87S/E89K)/Qβ RNA complex | |
| 2B2E | X-ray | CP(N87S/E89K)/MS2 RNA complex | |
| 2B2G | X-ray | CP(N87S)/MS2 RNA complex | |
| 2IZ8 | X-ray | CP/RNA complex, C-5 RNA operator with cytosine at -7 position | |
| 2IZ9 | X-ray | CP/RNA complex, RNA operator with pyridin-4-one at -5 position | |
| 2IZN | X-ray | CP/RNA complex, RNA operator with guanine at -10 position, and GC pairs above and below -10 position changed to CG | |
| 2IZM | X-ray | CP/RNA complex, C-5 RNA operator with cytosine at -10 position | |
| 2VTU | X-ray | Octahedral VLP (artifact) from covalent CP dimer fusion | |
| 4ZOR | X-ray | T = 1 VLP (S37P) | |
| 6RRS | Cryo-EM | T = 3 VLP | |
| 6RRT | Cryo-EM | T = 4 VLP | |
| Qβ | 1QBE | X-ray | T = 3 VLP |
| 5KIP | Cryo-EM | T = 3 VLP | |
| 5VLY | Cryo-EM | T = 3 VLP | |
| 5VLZ | Cryo-EM | Mature virion, capsid and Mat (A2) | |
| 5VM7 | Cryo-EM | MurA-A2 complex | |
| 7LGE | Cryo-EM | T = 4 VLP | |
| 7LGF | Cryo-EM | Prolate VLP | |
| 7LGG | Cryo-EM | Oblate VLP | |
| 7LGH | Cryo-EM | Small prolate VLP | |
| 7LHD | Cryo-EM | Complete atomic model of Qβ | |
| 7TJD | X-ray | T = 1 VLP | |
| PP7 | 6N4V | Cryo-EM | T = 4 VLP |
| 2QUX | X-ray | CP/RNA complex | |
| 2QUD | X-ray | CP dimer | |
| 1DWN | X-ray | T = 3 VLP | |
| AP205 | 5JZR | NMR | CP dimer |
| 5FS4 | X-ray | CP dimer | |
| 5LQP | Cryo-EM | T = 3 VLP | |
| ΦCb5 | 2W4Z | X-ray | T = 3 VLP, from mature virions |
| 2W4Y | X-ray | T = 3 VLP, from overexpression of CP | |
| PRR1 | 2VF9 | X-ray | T = 3 VLP |
| 4ANG | X-ray | T = 3 VLP with RNA operator | |
| fr | 1FRS | X-ray | T = 3 VLP, WT |
| 1FR5 | X-ray | T = 3 VLP, 4 residue deletion in FG loop | |
| GA | 1GAV | X-ray | T = 3 VLP |
| Nonculturable, pre-2018, ssRNA phages | |||
| AVE002 | 6YF9 | X-ray | T = 3 VLP |
| AVE015 | 6YFA | X-ray | T = 3 VLP |
| AVE016 | 6YFB | X-ray | Prolate VLP |
| AVE019 | 6YFC | X-ray | T = 3 VLP |
| Beihai14 | 6YFD | X-ray | T = 3 VLP, unusual CP |
| Beihai19 | 6YFE | X-ray | T = 3 VLP |
| Beihai21 | 6YFF | X-ray | T = 3 VLP |
| Beihai32 | 6YFG | X-ray | T = 3 VLP |
| EMS014 | 6YFH | X-ray | T = 3 VLP |
| 6YFI | X-ray | CP dimer | |
| ESE001 | 6YFJ | X-ray | T = 3 VLP |
| ESE007 | 6YFK | X-ray | T = 3 VLP |
| ESE020 | 6YFL | X-ray | T = 3 VLP |
| ESE021 | 6YFM | X-ray | T = 3 VLP |
| ESE058 | 6YFN | X-ray | T = 3 VLP |
| NT-214 | 6YFQ | X-ray | T = 3 VLP |
| NT-391 | 6YFR | X-ray | T = 3 VLP |
| Wenzhou1 | 6YFT | X-ray | T = 3 VLP |
| Wenzhou4 | 6YFU | X-ray | T = 3 VLP |
| PQ-465 | 6YFS | X-ray | T = 3 VLP |
| AC | 6YF7 | X-ray | T = 3 VLP |
| GQ-112 | 6YFP | X-ray | T = 3 VLP |
| GQ-907 | 6YFO | X-ray | T = 3 VLP |
References
- Loeb, T. Isolation of a bacteriophage specific for the F plus and Hfr mating types of Escherichia coli K-12. Science 1960, 131, 932–933. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, S.R.; Janowski, A.B.; Zhao, G.; Barouch, D.; Wang, D. Hyperexpansion of RNA Bacteriophage Diversity. PLoS Biol. 2016, 14, e1002409. [Google Scholar] [CrossRef] [PubMed]
- Callanan, J.; Stockdale, S.R.; Shkoporov, A.; Draper, L.A.; Ross, R.P.; Hill, C. Expansion of known ssRNA phage genomes: From tens to over a thousand. Sci. Adv. 2020, 6, eaay5981. [Google Scholar] [CrossRef] [PubMed]
- Neri, U.; Wolf, Y.I.; Roux, S.; Camargo, A.P.; Lee, B.; Kazlauskas, D.; Chen, I.M.; Ivanova, N.; Zeigler Allen, L.; Paez-Espino, D.; et al. Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell 2022, 185, 4023–4037.e18. [Google Scholar] [CrossRef]
- Wolf, Y.I.; Silas, S.; Wang, Y.; Wu, S.; Bocek, M.; Kazlauskas, D.; Krupovic, M.; Fire, A.; Dolja, V.V.; Koonin, E.V. Doubling of the known set of RNA viruses by metagenomic analysis of an aquatic virome. Nat. Microbiol. 2020, 5, 1262–1270. [Google Scholar] [CrossRef]
- Callanan, J.; Stockdale, S.R.; Adriaenssens, E.M.; Kuhn, J.H.; Rumnieks, J.; Pallen, M.J.; Shkoporov, A.N.; Draper, L.A.; Ross, R.P.; Hill, C. Leviviricetes: Expanding and restructuring the taxonomy of bacteria-infecting single-stranded RNA viruses. Microb. Genom. 2021, 7, 000686. [Google Scholar] [CrossRef]
- Epstein, S. National Academy of Sciences: Abstracts of Papers Presented at the Autumn Meeting, 29 October, La Jolla, California, 30 October-1 November 1961, Los Angeles. Science 1961, 134, 1425–1437. [Google Scholar] [CrossRef]
- Miyake, T.; Yanagisawa, K.; Watanabe, I. Alteration of a host character by infection with RNA phage Q-beta. Jpn. J. Microbiol. 1966, 10, 141–148. [Google Scholar] [CrossRef]
- Bradley, D.E. The Structure and Infective Process of a Pseudomonas Aeruginosa Bacteriophage Containing Ribonucleic Acid. Microbiology 1966, 45, 83–96. [Google Scholar] [CrossRef]
- Pourcel, C.; Midoux, C.; Vergnaud, G.; Latino, L. A carrier state is established in Pseudomonas aeruginosa by phage LeviOr01, a newly isolated ssRNA levivirus. J. Gen. Virol. 2017, 98, 2181–2189. [Google Scholar] [CrossRef]
- Coffi, H. Lysotypie des Acinetobacter. Master’s Thesis, Laval University, Québec, QC, Canada, 1995. [Google Scholar]
- Schmidt, J.M.; Stanier, R.Y. Isolation and Characterization of Bacteriophages Active against Stalked Bacteria. J. Gen. Microbiol. 1965, 39, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Olsen, R.H.; Thomas, D.D. Characteristics and purification of PRR1, an RNA phage specific for the broad host range Pseudomonas R1822 drug resistance plasmid. J. Virol. 1973, 12, 1560–1567. [Google Scholar] [CrossRef] [PubMed]
- Coetzee, J.N.; Bradley, D.E.; Hedges, R.W.; Fleming, J.; Lecatsas, G. Bacteriophage M: An incompatibility group M plasmid-specific phage. J. Gen. Microbiol. 1983, 129, 2271–2276. [Google Scholar] [CrossRef] [PubMed]
- Sirgel, F.A.; Coetzee, J.N.; Hedges, R.W.; Lecatsas, G. Phage C-1: An IncC group; plasmid-specific phage. J. Gen. Microbiol. 1981, 122, 155–160. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, D.; Maker, D.; Colleran, E. A method for the direct isolation of IncH plasmid-dependent bacteriophages. Lett. Appl. Microbiol. 1987, 5, 37–40. [Google Scholar] [CrossRef]
- Carmichael, G.G.; Weber, K.; Niveleau, A.; Wahba, A.J. The host factor required for RNA phage Qbeta RNA replication in vitro. Intracellular location, quantitation, and purification by polyadenylate-cellulose chromatography. J. Biol. Chem. 1975, 250, 3607–3612. [Google Scholar] [CrossRef]
- Blumenthal, T.; Landers, T.A.; Weber, K. Bacteriophage Q replicase contains the protein biosynthesis elongation factors EF Tu and EF Ts. Proc. Natl. Acad. Sci. USA 1972, 69, 1313–1317. [Google Scholar] [CrossRef]
- Rumnieks, J.; Lieknina, I.; Kalnins, G.; Sisovs, M.; Akopjana, I.; Bogans, J.; Tars, K. Three-dimensional structure of 22 uncultured ssRNA bacteriophages: Flexibility of the coat protein fold and variations in particle shapes. Sci. Adv. 2020, 6, eabc0023. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, Z.; Lai, M.; Shu, S.; Du, Y.; Zhou, Z.H.; Sun, R. In situ structures of the genome and genome-delivery apparatus in a single-stranded RNA virus. Nature 2017, 541, 112–116. [Google Scholar] [CrossRef]
- Gorzelnik, K.V.; Cui, Z.; Reed, C.A.; Jakana, J.; Young, R.; Zhang, J. Asymmetric cryo-EM structure of the canonical Allolevivirus Qbeta reveals a single maturation protein and the genomic ssRNA in situ. Proc. Natl. Acad. Sci. USA 2016, 113, 11519–11524. [Google Scholar] [CrossRef]
- Koning, R.I.; Gomez-Blanco, J.; Akopjana, I.; Vargas, J.; Kazaks, A.; Tars, K.; Carazo, J.M.; Koster, A.J. Asymmetric cryo-EM reconstruction of phage MS2 reveals genome structure in situ. Nat. Commun. 2016, 7, 12524. [Google Scholar] [CrossRef]
- Zhong, Q.; Carratala, A.; Nazarov, S.; Guerrero-Ferreira, R.C.; Piccinini, L.; Bachmann, V.; Leiman, P.G.; Kohn, T. Genetic, Structural, and Phenotypic Properties of MS2 Coliphage with Resistance to ClO(2) Disinfection. Environ. Sci. Technol. 2016, 50, 13520–13528. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.; Gorzelnik, K.V.; Chang, J.Y.; Langlais, C.; Jakana, J.; Young, R.; Zhang, J. Structures of Qbeta virions, virus-like particles, and the Qbeta-MurA complex reveal internal coat proteins and the mechanism of host lysis. Proc. Natl. Acad. Sci. USA 2017, 114, 11697–11702. [Google Scholar] [CrossRef]
- Steitz, J.A. Identification of the A protein as a structural component of bacteriophage R17. J. Mol. Biol. 1968, 33, 923–936. [Google Scholar] [CrossRef]
- Steitz, J.A. Isolation of the A protein from bacteriphage R17. J. Mol. Biol. 1968, 33, 937–945. [Google Scholar] [CrossRef]
- Reed, C.A.; Langlais, C.; Kuznetsov, V.; Young, R. Inhibitory mechanism of the Qbeta lysis protein A2. Mol. Microbiol. 2012, 86, 836–844. [Google Scholar] [CrossRef]
- Rumnieks, J.; Tars, K. Crystal Structure of the Maturation Protein from Bacteriophage Qbeta. J. Mol. Biol. 2017, 429, 688–696. [Google Scholar] [CrossRef]
- Chang, J.Y.; Cui, Z.; Yang, K.; Huang, J.; Minary, P.; Zhang, J. Hierarchical natural move Monte Carlo refines flexible RNA structures into cryo-EM densities. RNA 2020, 26, 1755–1766. [Google Scholar] [CrossRef] [PubMed]
- Manchak, J.; Anthony, K.G.; Frost, L.S. Mutational analysis of F-pilin reveals domains for pilus assembly, phage infection and DNA transfer. Mol. Microbiol. 2002, 43, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Meng, R.; Jiang, M.; Cui, Z.; Chang, J.Y.; Yang, K.; Jakana, J.; Yu, X.; Wang, Z.; Hu, B.; Zhang, J. Structural basis for the adsorption of a single-stranded RNA bacteriophage. Nat. Commun. 2019, 10, 3130. [Google Scholar] [CrossRef]
- Bradley, D.E. Shortening of Pseudomonas aeruginosa pili after RNA-phage adsorption. J. Gen. Microbiol. 1972, 72, 303–319. [Google Scholar] [CrossRef]
- Bayer, M.; Eferl, R.; Zellnig, G.; Teferle, K.; Dijkstra, A.; Koraimann, G.; Hogenauer, G. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J. Bacteriol. 1995, 177, 4279–4288. [Google Scholar] [CrossRef]
- Paranchych, W.; Ainsworth, S.K.; Dick, A.J.; Krahn, P.M. Stages in phage R17 infection. V. Phage eclipse and the role of F pili. Virology 1971, 45, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Danziger, R.E.; Paranchych, W. Stages in phage R17 infection. 3. Energy requirements for the F-pili mediated eclipse of viral infectivity. Virology 1970, 40, 554–564. [Google Scholar] [CrossRef] [PubMed]
- Dent, K.C.; Thompson, R.; Barker, A.M.; Hiscox, J.A.; Barr, J.N.; Stockley, P.G.; Ranson, N.A. The asymmetric structure of an icosahedral virus bound to its receptor suggests a mechanism for genome release. Structure 2013, 21, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Harb, L.; Chamakura, K.; Khara, P.; Christie, P.J.; Young, R.; Zeng, L. ssRNA phage penetration triggers detachment of the F-pilus. Proc. Natl. Acad. Sci. USA 2020, 117, 25751–25758. [Google Scholar] [CrossRef] [PubMed]
- Schoulaker, R.; Engelberg-Kulka, H. Escherichia coli mutant temperature sensitive for group I RNA bacteriophages. J. Virol. 1978, 25, 433–435. [Google Scholar] [CrossRef] [PubMed]
- Gytz, H.; Mohr, D.; Seweryn, P.; Yoshimura, Y.; Kutlubaeva, Z.; Dolman, F.; Chelchessa, B.; Chetverin, A.B.; Mulder, F.A.; Brodersen, D.E.; et al. Structural basis for RNA-genome recognition during bacteriophage Qbeta replication. Nucleic Acids Res. 2015, 43, 10893–10906. [Google Scholar] [CrossRef]
- Kawashima, T.; Berthet-Colominas, C.; Wulff, M.; Cusack, S.; Leberman, R. The structure of the Escherichia coli EF-Tu.EF-Ts complex at 2.5 A resolution. Nature 1996, 379, 511–518. [Google Scholar] [CrossRef]
- Sengupta, J.; Agrawal, R.K.; Frank, J. Visualization of protein S1 within the 30S ribosomal subunit and its interaction with messenger RNA. Proc. Natl. Acad. Sci. USA 2001, 98, 11991–11996. [Google Scholar] [CrossRef]
- Kita, H.; Cho, J.; Matsuura, T.; Nakaishi, T.; Taniguchi, I.; Ichikawa, T.; Shima, Y.; Urabe, I.; Yomo, T. Functional Qbeta replicase genetically fusing essential subunits EF-Ts and EF-Tu with beta-subunit. J. Biosci. Bioeng. 2006, 101, 421–426. [Google Scholar] [CrossRef]
- Takeshita, D.; Tomita, K. Assembly of Qbeta viral RNA polymerase with host translational elongation factors EF-Tu and -Ts. Proc. Natl. Acad. Sci. USA 2010, 107, 15733–15738. [Google Scholar] [CrossRef]
- Kidmose, R.T.; Vasiliev, N.N.; Chetverin, A.B.; Andersen, G.R.; Knudsen, C.R. Structure of the Qbeta replicase, an RNA-dependent RNA polymerase consisting of viral and host proteins. Proc. Natl. Acad. Sci. USA 2010, 107, 10884–10889. [Google Scholar] [CrossRef]
- Takeshita, D.; Tomita, K. Molecular basis for RNA polymerization by Qbeta replicase. Nat. Struct. Mol. Biol. 2012, 19, 229–237. [Google Scholar] [CrossRef]
- Kutlubaeva, Z.S.; Chetverina, H.V.; Chetverin, A.B. The Contribution of Ribosomal Protein S1 to the Structure and Function of Qbeta Replicase. Acta Naturae 2017, 9, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Pei, J.; Grishin, N.V. AL2CO: Calculation of positional conservation in a protein sequence alignment. Bioinformatics 2001, 17, 700–712. [Google Scholar] [CrossRef] [PubMed]
- Miranda, G.; Schuppli, D.; Barrera, I.; Hausherr, C.; Sogo, J.M.; Weber, H. Recognition of bacteriophage Qbeta plus strand RNA as a template by Qbeta replicase: Role of RNA interactions mediated by ribosomal proteins S1 and host factor. J. Mol. Biol. 1997, 267, 1089–1103. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, D.; Yamashita, S.; Tomita, K. Molecular insights into replication initiation by Qbeta replicase using ribosomal protein S1. Nucleic Acids Res. 2014, 42, 10809–10822. [Google Scholar] [CrossRef]
- Takeshita, D.; Yamashita, S.; Tomita, K. Mechanism for template-independent terminal adenylation activity of Qbeta replicase. Structure 2012, 20, 1661–1669. [Google Scholar] [CrossRef]
- Tomita, K. Structures and functions of Qbeta replicase: Translation factors beyond protein synthesis. Int. J. Mol. Sci. 2014, 15, 15552–15570. [Google Scholar] [CrossRef] [PubMed]
- Vasilyev, N.N.; Kutlubaeva, Z.S.; Ugarov, V.I.; Chetverina, H.V.; Chetverin, A.B. Ribosomal protein S1 functions as a termination factor in RNA synthesis by Qbeta phage replicase. Nat. Commun. 2013, 4, 1781. [Google Scholar] [CrossRef]
- Hohn, T. Packaging of Genomes in Bacteriophages: A Comparison of ssRNA Bacteriophages and dsDNA Bactgeriophages. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1976, 276, 143–150. [Google Scholar] [CrossRef]
- de Haas, F.; Paatero, A.O.; Mindich, L.; Bamford, D.H.; Fuller, S.D. A symmetry mismatch at the site of RNA packaging in the polymerase complex of dsRNA bacteriophage phi6. J. Mol. Biol. 1999, 294, 357–372. [Google Scholar] [CrossRef]
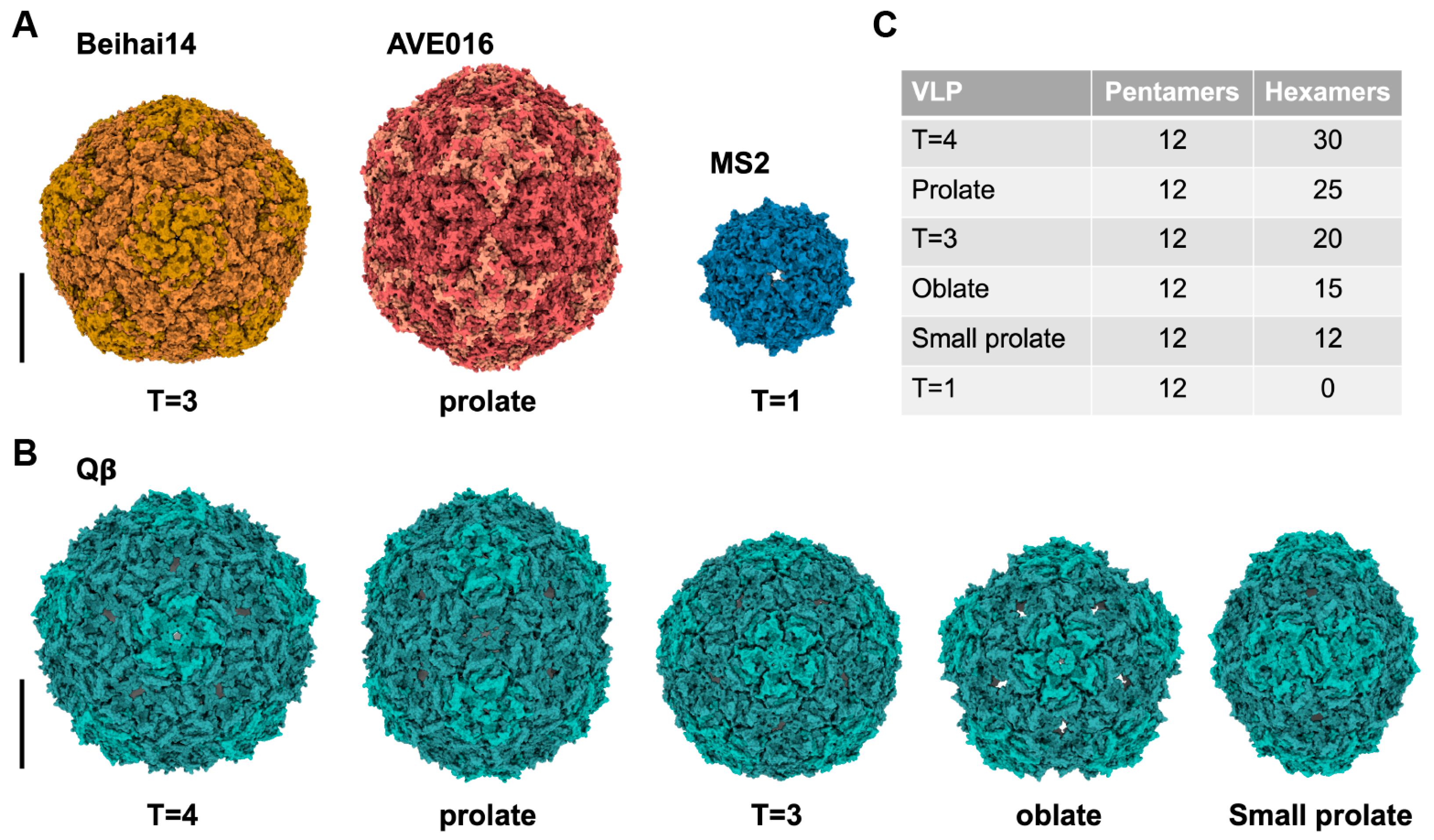
- Chang, J.Y.; Gorzelnik, K.V.; Thongchol, J.; Zhang, J. Structural Assembly of Qbeta Virion and Its Diverse Forms of Virus-like Particles. Viruses 2022, 14, 225. [Google Scholar] [CrossRef] [PubMed]
- Fouts, D.E.; True, H.L.; Celander, D.W. Functional recognition of fragmented operator sites by R17/MS2 coat protein, a translational repressor. Nucleic Acids Res. 1997, 25, 4464–4473. [Google Scholar] [CrossRef] [PubMed]
- Lim, F.; Spingola, M.; Peabody, D.S. The RNA-binding site of bacteriophage Qbeta coat protein. J. Biol. Chem. 1996, 271, 31839–31845. [Google Scholar] [CrossRef] [PubMed]
- Persson, M.; Tars, K.; Liljas, L. PRR1 coat protein binding to its RNA translational operator. Acta Crystallogr. Sect. D Biol. Crystallogr. 2013, 69, 367–372. [Google Scholar] [CrossRef]
- Comas-Garcia, M. Packaging of Genomic RNA in Positive-Sense Single-Stranded RNA Viruses: A Complex Story. Viruses 2019, 11, 253. [Google Scholar] [CrossRef]
- de Martin Garrido, N.; Crone, M.A.; Ramlaul, K.; Simpson, P.A.; Freemont, P.S.; Aylett, C.H.S. Bacteriophage MS2 displays unreported capsid variability assembling T = 4 and mixed capsids. Mol. Microbiol. 2020, 113, 143–152. [Google Scholar] [CrossRef]
- Zhao, L.; Kopylov, M.; Potter, C.S.; Carragher, B.; Finn, M.G. Engineering the PP7 Virus Capsid as a Peptide Display Platform. ACS Nano 2019, 13, 4443–4454. [Google Scholar] [CrossRef] [PubMed]
- Lieknina, I.; Kalnins, G.; Akopjana, I.; Bogans, J.; Sisovs, M.; Jansons, J.; Rumnieks, J.; Tars, K. Production and characterization of novel ssRNA bacteriophage virus-like particles from metagenomic sequencing data. J. Nanobiotechnol. 2019, 17, 61. [Google Scholar] [CrossRef]
- Asensio, M.A.; Morella, N.M.; Jakobson, C.M.; Hartman, E.C.; Glasgow, J.E.; Sankaran, B.; Zwart, P.H.; Tullman-Ercek, D. A Selection for Assembly Reveals That a Single Amino Acid Mutant of the Bacteriophage MS2 Coat Protein Forms a Smaller Virus-like Particle. Nano Lett. 2016, 16, 5944–5950. [Google Scholar] [CrossRef] [PubMed]
- Cielens, I.; Ose, V.; Petrovskis, I.; Strelnikova, A.; Renhofa, R.; Kozlovska, T.; Pumpens, P. Mutilation of RNA phage Qbeta virus-like particles: From icosahedrons to rods. FEBS Lett. 2000, 482, 261–264. [Google Scholar] [CrossRef]
- Young, R. Phage lysis: Three steps, three choices, one outcome. J. Microbiol. 2014, 52, 243–258. [Google Scholar] [CrossRef]
- Chamakura, K.R.; Young, R. Single-gene lysis in the metagenomic era. Curr. Opin. Microbiol. 2020, 56, 109–117. [Google Scholar] [CrossRef]
- Chamakura, K.R.; Edwards, G.B.; Young, R. Mutational analysis of the MS2 lysis protein L. Microbiology 2017, 163, 961–969. [Google Scholar] [CrossRef]
- Reed, C.A.; Langlais, C.; Wang, I.N.; Young, R. A(2) expression and assembly regulates lysis in Qbeta infections. Microbiology 2013, 159, 507–514. [Google Scholar] [CrossRef]
- Chamakura, K.R.; Sham, L.T.; Davis, R.M.; Min, L.; Cho, H.; Ruiz, N.; Bernhardt, T.G.; Young, R. A viral protein antibiotic inhibits lipid II flippase activity. Nat. Microbiol. 2017, 2, 1480–1484. [Google Scholar] [CrossRef]
- Adler, B.A.; Chamakura, K.; Carion, H.; Krog, J.; Deutschbauer, A.M.; Young, R.; Mutalik, V.K.; Arkin, A.P. Multicopy suppressor screens reveal convergent evolution of single-gene lysis proteins. Nat. Chem. Biol. 2023, 19, 759–766. [Google Scholar] [CrossRef] [PubMed]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Baek, M.; DiMaio, F.; Anishchenko, I.; Dauparas, J.; Ovchinnikov, S.; Lee, G.R.; Wang, J.; Cong, Q.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef] [PubMed]
- Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Bell, E.W.; Zhang, Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep. Methods 2021, 1, 100014. [Google Scholar] [CrossRef] [PubMed]
- Lin, Z.; Akin, H.; Rao, R.; Hie, B.; Zhu, Z.; Lu, W.; Smetanin, N.; Verkuil, R.; Kabeli, O.; Shmueli, Y.; et al. Evolutionary-scale prediction of atomic-level protein structure with a language model. Science 2023, 379, 1123–1130. [Google Scholar] [CrossRef] [PubMed]
- Amann, S.J.; Keihsler, D.; Bodrug, T.; Brown, N.G.; Haselbach, D. Frozen in time: Analyzing molecular dynamics with time-resolved cryo-EM. Structure 2023, 31, 4–19. [Google Scholar] [CrossRef]
- Wagner, F.R.; Watanabe, R.; Schampers, R.; Singh, D.; Persoon, H.; Schaffer, M.; Fruhstorfer, P.; Plitzko, J.; Villa, E. Preparing samples from whole cells using focused-ion-beam milling for cryo-electron tomography. Nat. Protoc. 2020, 15, 2041–2070. [Google Scholar] [CrossRef]
- Franze de Fernandez, M.T.; Eoyang, L.; August, J.T. Factor fraction required for the synthesis of bacteriophage Qbeta-RNA. Nature 1968, 219, 588–590. [Google Scholar] [CrossRef]
- Franze de Fernandez, M.T.; Hayward, W.S.; August, J.T. Bacterial proteins required for replication of phage Q ribonucleic acid. Pruification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 1972, 247, 824–831. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Weise, L.I.; Mutschler, H. In vitro characterisation of the MS2 RNA polymerase complex reveals host factors that modulate emesviral replicase activity. Commun. Biol. 2022, 5, 264. [Google Scholar] [CrossRef]


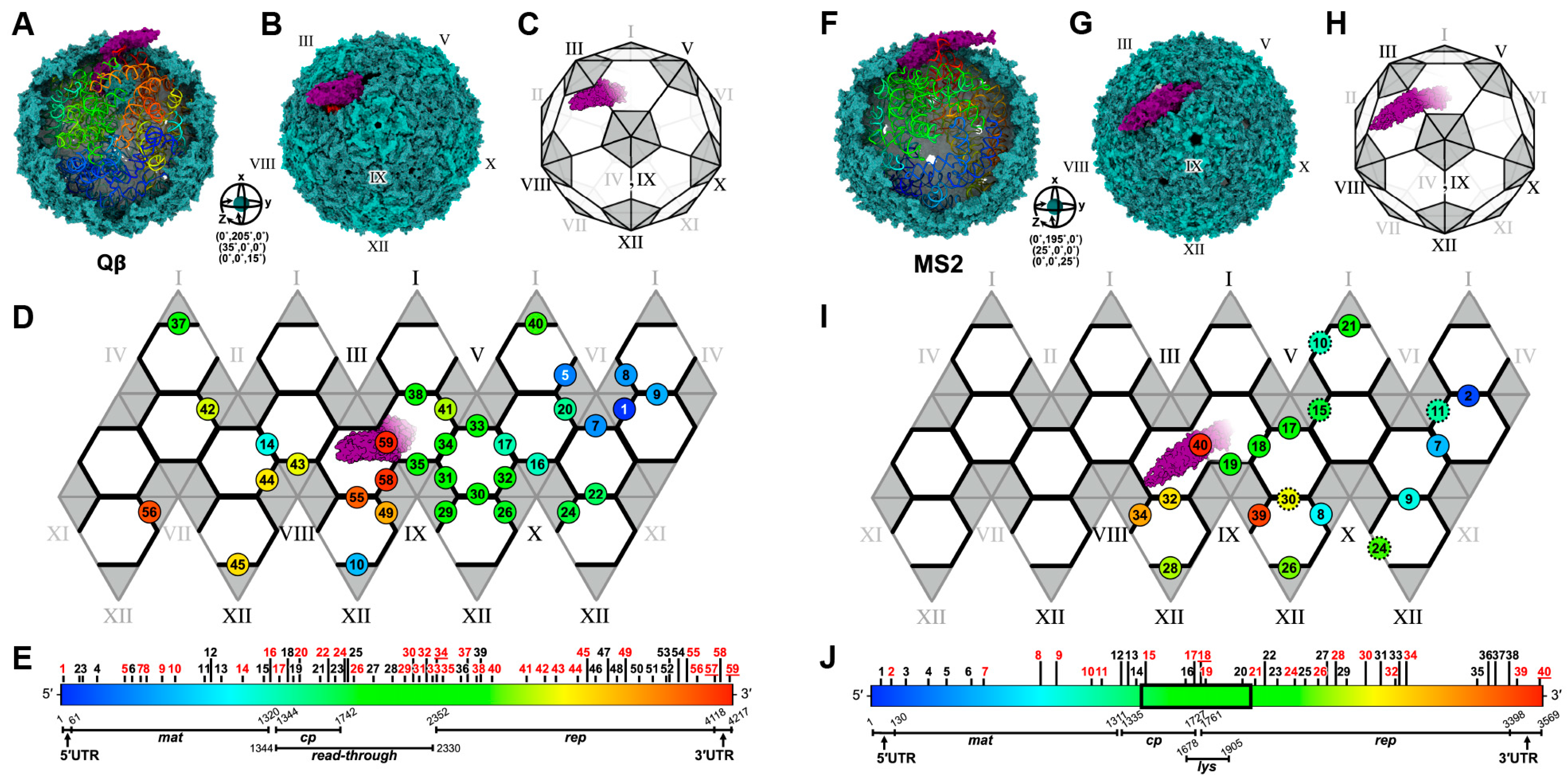

| Phages | Total Particles | Mature Virion Particles * | Reference |
|---|---|---|---|
| MS2 | 22,441 | 18,977 (~85%) | Konning R.I., 2016 [22] |
| 48,276 | 44,897 (~93%) | Meng R., 2019 [31] | |
| Qβ | 51,815 | 12,975 (~25%) | Gorzelnik K.V., 2016 [21] |
| 248,445 | 76,843 (~31%) | Cui Z., 2017 [24] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thongchol, J.; Lill, Z.; Hoover, Z.; Zhang, J. Recent Advances in Structural Studies of Single-Stranded RNA Bacteriophages. Viruses 2023, 15, 1985. https://doi.org/10.3390/v15101985
Thongchol J, Lill Z, Hoover Z, Zhang J. Recent Advances in Structural Studies of Single-Stranded RNA Bacteriophages. Viruses. 2023; 15(10):1985. https://doi.org/10.3390/v15101985
Chicago/Turabian StyleThongchol, Jirapat, Zachary Lill, Zachary Hoover, and Junjie Zhang. 2023. "Recent Advances in Structural Studies of Single-Stranded RNA Bacteriophages" Viruses 15, no. 10: 1985. https://doi.org/10.3390/v15101985





