Electrostatic Surface Potential as a Key Parameter in Virus Transmission and Evolution: How to Manage Future Virus Pandemics in the Post-COVID-19 Era
Abstract
:1. Introduction
2. Definition of the Electrostatic Surface Potential
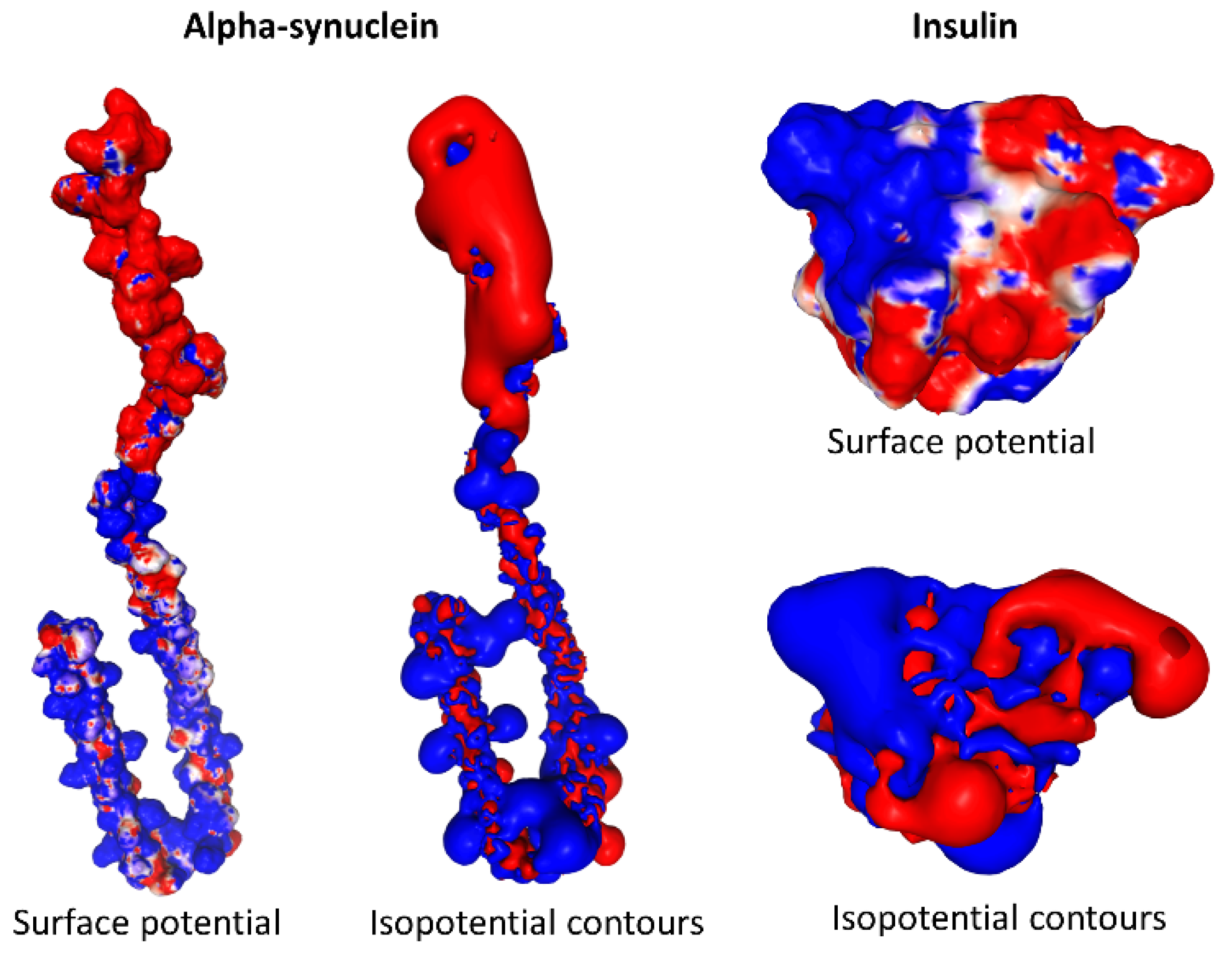
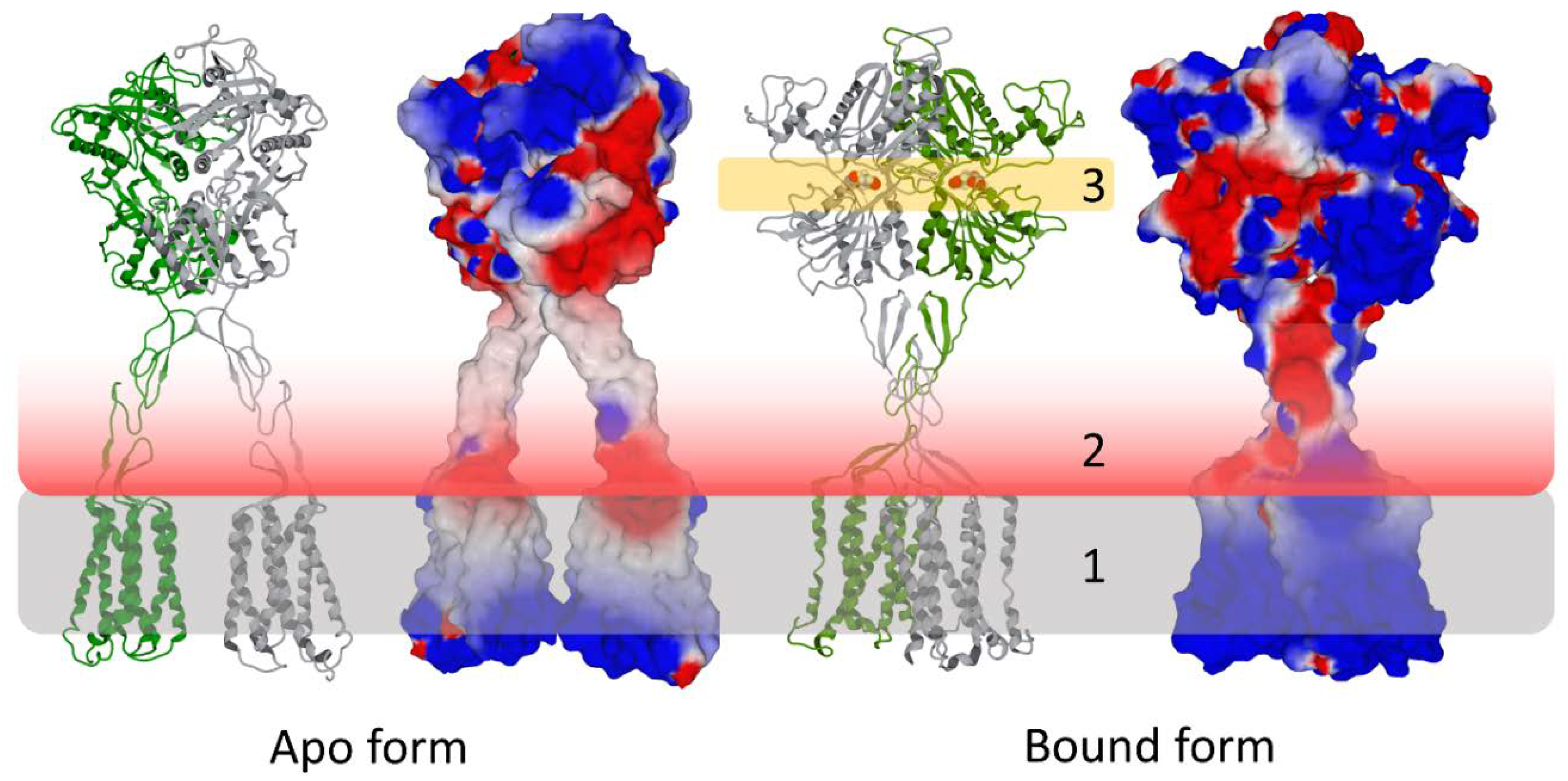
3. Biological Significance of the Electrostatic Surface Potential
4. Electrostatic Surface Potential in HIV-1 Evolution
5. Electrostatic Surface Potential in Influenza Virus Evolution
6. Electrostatic Surface Potential in Coronavirus Evolution
7. Clues for Managing Future Pandemics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fantini, J.; Yahi, N. Brain Lipids in Synaptic Function and Neurological Disease: Clues to Innovative Therapeutic Strategies for Brain Disorders; Academic Press: Cambridge, MA, USA, 2015. [Google Scholar]
- Chaplin, M. Do we underestimate the importance of water in cell biology? Nat. Rev. Mol. Cell Biol. 2006, 7, 861–866. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Wikswo, J. Dimensions of systems biology. Rev. Physiol. Biochem. Pharmacol. 2006, 157, 81–104. [Google Scholar] [PubMed]
- Brodsky, W.Y. Protein synthesis rhythm. J. Theor. Biol. 1975, 55, 167–200. [Google Scholar] [CrossRef] [PubMed]
- McFadden, J.; Al-Khalili, J. The origins of quantum biology. Proc. R. Soc. A 2018, 474, 20180674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weiner, P.K.; Langridge, R.; Blaney, J.M.; Schaefer, R.; Kollman, P.A. Electrostatic potential molecular surfaces. Proc. Natl. Acad. Sci. USA 1982, 79, 3754–3758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fantini, J.; Yahi, N.; Azzaz, F.; Chahinian, H. Structural dynamics of SARS-CoV-2 variants: A health monitoring strategy for anticipating Covid-19 outbreaks. J. Infect. 2021, 83, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Repits, J.; Sterjovski, J.; Badia-Martinez, D.; Mild, M.; Gray, L.; Churchill, M.J.; Purcell, D.F.; Karlsson, A.; Albert, J.; Fenyö, E.M. Primary HIV-1 R5 isolates from end-stage disease display enhanced viral fitness in parallel with increased gp120 net charge. Virology 2008, 379, 125–134. [Google Scholar] [CrossRef] [Green Version]
- Heidari, A.; Righetto, I.; Filippini, F. Electrostatic variation of haemagglutinin as a hallmark of the evolution of avian influenza viruses. Sci. Rep. 2018, 8, 1929. [Google Scholar] [CrossRef] [Green Version]
- Kalinina, O.V.; Pfeifer, N.; Lengauer, T. Modelling binding between CCR5 and CXCR4 receptors and their ligands suggests the surface electrostatic potential of the co-receptor to be a key player in the HIV-1 tropism. Retrovirology 2013, 10, 130. [Google Scholar] [CrossRef] [Green Version]
- Honig, B.; Nicholls, A. Classical electrostatics in biology and chemistry. Science 1995, 268, 1144–1149. [Google Scholar] [CrossRef]
- López de Victoria, A.; Kieslich, C.A.; Rizos, A.K.; Krambovitis, E.; Morikis, D. Clustering of HIV-1 subtypes based on gp120 V3 loop electrostatic properties. BMC Biophys. 2012, 5, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fogolari, F.; Brigo, A.; Molinari, H. The Poisson–Boltzmann equation for biomolecular electrostatics: A tool for structural biology. J. Mol. Recognit. 2002, 15, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Sharp, K.A.; Honig, B. Calculating total electrostatic energies with the nonlinear Poisson-Boltzmann equation. J. Phys. Chem. 1990, 94, 7684–7692. [Google Scholar] [CrossRef]
- Murray, J.S.; Politzer, P. The electrostatic potential: An overview. Wiley Interdiscip. Rev. Comput. Mol. Sci. 2011, 1, 153–163. [Google Scholar] [CrossRef]
- Jo, S.; Vargyas, M.; Vasko-Szedlar, J.; Roux, B.; Im, W. PBEQ-Solver for online visualization of electrostatic potential of biomolecules. Nucleic Acids Res. 2008, 36, W270–W275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Im, W.; Beglov, D.; Roux, B. Continuum solvation model: Computation of electrostatic forces from numerical solutions to the Poisson-Boltzmann equation. Comput. Phys. Commun. 1998, 111, 59–75. [Google Scholar] [CrossRef]
- Auffinger, P.; Hashem, Y. Nucleic acid solvation: From outside to insight. Curr. Opin. Struct. Biol. 2007, 17, 325–333. [Google Scholar] [CrossRef]
- Fantini, J.; Garmy, N.; Mahfoud, R.; Yahi, N. Lipid rafts: Structure, function and role in HIV, Alzheimer’s and prion diseases. Expert Rev. Mol. Med. 2002, 4, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Fantini, J.; Chahinian, H.; Yahi, N. Progress toward Alzheimer’s disease treatment: Leveraging the Achilles’ heel of Aβ oligomers? Protein Sci. A Publ. Protein Soc. 2020, 29, 1748–1759. [Google Scholar] [CrossRef]
- Fontes, A.; Fernandes, H.; Barjas-Castro, M.; de Thomaz, A.; de Ysasa Pozzo, L.; Barbosa, L.; Cesar, C. Red Blood Cell Membrane Viscoelasticity, Agglutination, and Zeta Potential Measurements with Double Optical Tweezers; SPIE: Bellingham, WA, USA, 2006; Volume 6088. [Google Scholar]
- Luner, S.J.; Sturgeon, P.; Szklarek, D.; McQuiston, D.T. Effects of Proteases and Neuraminidase on RBC Surface Charge and Agglutination: A Klinetic Study 1. Vox Sang. 1975, 28, 184–199. [Google Scholar] [CrossRef]
- Van Oss, C.; Absolom, D. Hemagglutination and the closest distance of approach of normal, neuraminidase-and papain-treated erythrocytes. Vox Sang. 1984, 47, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Schnaar, R.L. Gangliosides of the Vertebrate Nervous System. J. Mol. Biol. 2016, 428, 3325–3336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schnaar, R.L.; Gerardy-Schahn, R.; Hildebrandt, H. Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014, 94, 461–518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azzaz, F.; Yahi, N.; Di Scala, C.; Chahinian, H.; Fantini, J. Ganglioside binding domains in proteins: Physiological and pathological mechanisms. Adv. Protein Chem. Struct. Biol. 2022, 128, 289–324. [Google Scholar] [CrossRef] [PubMed]
- Koehl, A.; Hu, H.; Feng, D.; Sun, B.; Zhang, Y.; Robertson, M.J.; Chu, M.; Kobilka, T.S.; Laeremans, T.; Steyaert, J.; et al. Structural insights into the activation of metabotropic glutamate receptors. Nature 2019, 566, 79–84. [Google Scholar] [CrossRef]
- Amin, M.; Küpper, J. Variations in proteins dielectric constants. ChemistryOpen 2020, 9, 691–694. [Google Scholar] [CrossRef]
- Mañes, S.; del Real, G.; Lacalle, R.A.; Lucas, P.; Gómez-Moutón, C.; Sánchez-Palomino, S.; Delgado, R.; Alcamí, J.; Mira, E.; Martínez, A.C. Membrane raft microdomains mediate lateral assemblies required for HIV-1 infection. EMBO Rep 2000, 1, 190–196. [Google Scholar] [CrossRef]
- Mañes, S.; del Real, G.; Martínez, A.C. Pathogens: Raft hijackers. Nat. Rev. Immunol. 2003, 3, 557–568. [Google Scholar] [CrossRef]
- Ripa, I.; Andreu, S.; López-Guerrero, J.A.; Bello-Morales, R. Membrane rafts: Portals for viral entry. Front. Microbiol. 2021, 12, 631274. [Google Scholar] [CrossRef]
- Omasta, B.; Tomaskova, J. Cellular Lipids—Hijacked Victims of Viruses. Viruses 2022, 14, 1896. [Google Scholar] [CrossRef]
- Pike, L.J. Lipid rafts: Bringing order to chaos. J. Lipid Res. 2003, 44, 655–667. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, Y. Gangliosides as influenza virus receptors. Variation of influenza viruses and their recognition of the receptor sialo-sugar chains. Prog. Lipid Res. 1994, 33, 429–457. Prog. Lipid Res. 1994, 33, 429–457. [Google Scholar] [CrossRef]
- Markwell, M.; Svennerholm, L.; Paulson, J.C. Specific gangliosides function as host cell receptors for Sendai virus. Proc. Natl. Acad. Sci. USA 1981, 78, 5406–5410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campanero-Rhodes, M.A.; Smith, A.; Chai, W.; Sonnino, S.; Mauri, L.; Childs, R.A.; Zhang, Y.; Ewers, H.; Helenius, A.; Imberty, A. N-glycolyl GM1 ganglioside as a receptor for simian virus 40. J. Virol. 2007, 81, 12846–12858. [Google Scholar] [CrossRef] [Green Version]
- Maginnis, M.S. Virus–receptor interactions: The key to cellular invasion. J. Mol. Biol. 2018, 430, 2590–2611. [Google Scholar] [CrossRef] [PubMed]
- Rolsma, M.D.; Kuhlenschmidt, T.B.; Gelberg, H.B.; Kuhlenschmidt, M.S. Structure and function of a ganglioside receptor for porcine rotavirus. J. Virol. 1998, 72, 9079–9091. [Google Scholar] [CrossRef] [Green Version]
- Hammache, D.; Piéroni, G.; Yahi, N.; Delézay, O.; Koch, N.; Lafont, H.; Tamalet, C.; Fantini, J. Specific interaction of HIV-1 and HIV-2 surface envelope glycoproteins with monolayers of galactosylceramide and ganglioside GM3. J. Biol. Chem. 1998, 273, 7967–7971. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammache, D.; Yahi, N.; Maresca, M.; Piéroni, G.; Fantini, J. Human erythrocyte glycosphingolipids as alternative cofactors for human immunodeficiency virus type 1 (HIV-1) entry: Evidence for CD4-induced interactions between HIV-1 gp120 and reconstituted membrane microdomains of glycosphingolipids (Gb3 and GM3). J. Virol. 1999, 73, 5244–5248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammache, D.; Yahi, N.; Piéroni, G.; Ariasi, F.; Tamalet, C.; Fantini, J. Sequential interaction of CD4 and HIV-1 gp120 with a reconstituted membrane patch of ganglioside GM3: Implications for the role of glycolipids as potential HIV-1 fusion cofactors. Biochem. Biophys. Res. Commun. 1998, 246, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Hug, P.; Lin, H.M.; Korte, T.; Xiao, X.; Dimitrov, D.S.; Wang, J.M.; Puri, A.; Blumenthal, R. Glycosphingolipids promote entry of a broad range of human immunodeficiency virus type 1 isolates into cell lines expressing CD4, CXCR4, and/or CCR5. J. Virol. 2000, 74, 6377–6385. [Google Scholar] [CrossRef]
- Harouse, J.M.; Bhat, S.; Spitalnik, S.L.; Laughlin, M.; Stefano, K.; Silberberg, D.H.; Gonzalez-Scarano, F. Inhibition of entry of HIV-1 in neural cell lines by antibodies against galactosyl ceramide. Science 1991, 253, 320–323. [Google Scholar] [CrossRef] [PubMed]
- Yahi, N.; Baghdiguian, S.; Moreau, H.; Fantini, J. Galactosyl ceramide (or a closely related molecule) is the receptor for human immunodeficiency virus type 1 on human colon epithelial HT29 cells. J. Virol. 1992, 66, 4848–4854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fantini, J.; Garmy, N.; Yahi, N. Prediction of glycolipid-binding domains from the amino acid sequence of lipid raft-associated proteins: Application to HpaA, a protein involved in the adhesion of Helicobacter pylori to gastrointestinal cells. Biochemistry 2006, 45, 10957–10962. [Google Scholar] [CrossRef] [PubMed]
- Mahfoud, R.; Garmy, N.; Maresca, M.; Yahi, N.; Puigserver, A.; Fantini, J. Identification of a common sphingolipid-binding domain in Alzheimer, prion, and HIV-1 proteins. J. Biol. Chem. 2002, 277, 11292–11296. [Google Scholar] [CrossRef] [Green Version]
- Fantini, J.; Chahinian, H.; Yahi, N. Leveraging coronavirus binding to gangliosides for innovative vaccine and therapeutic strategies against COVID-19. Biochem. Biophys. Res. Commun. 2021, 538, 132–136. [Google Scholar] [CrossRef]
- Fantini, J.; Chahinian, H.; Yahi, N. A Vaccine Strategy Based on the Identification of an Annular Ganglioside Binding Motif in Monkeypox Virus Protein E8L. Viruses 2022, 14, 2531. [Google Scholar] [CrossRef]
- Fantini, J. How sphingolipids bind and shape proteins: Molecular basis of lipid-protein interactions in lipid shells, rafts and related biomembrane domains. Cell. Mol. Life Sci. CMLS 2003, 60, 1027–1032. [Google Scholar] [CrossRef]
- Froimowitz, M. HyperChem: A software package for computational chemistry and molecular modeling. BioTechniques 1993, 14, 1010–1013. [Google Scholar]
- Bour, S.; Geleziunas, R.; Wainberg, M.A. The human immunodeficiency virus type 1 (HIV-1) CD4 receptor and its central role in promotion of HIV-1 infection. Microbiol. Rev. 1995, 59, 63–93. [Google Scholar] [CrossRef]
- Moore, J.P.; Trkola, A.; Dragic, T. Co-receptors for HIV-1 entry. Curr. Opin. Immunol. 1997, 9, 551–562. [Google Scholar] [CrossRef]
- Moore, J.P.; Kitchen, S.G.; Pugach, P.; Zack, J.A. The CCR5 and CXCR4 coreceptors—Central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retrovir. 2004, 20, 111–126. [Google Scholar] [CrossRef] [PubMed]
- Regoes, R.R.; Bonhoeffer, S. The HIV coreceptor switch: A population dynamical perspective. Trends Microbiol. 2005, 13, 269–277. [Google Scholar] [CrossRef] [PubMed]
- Briggs, D.R.; Tuttle, D.L.; Sleasman, J.W.; Goodenow, M.M. Envelope V3 amino acid sequence predicts HIV-1 phenotype (co-receptor usage and tropism for macrophages). AIDS 2000, 14, 2937–2939. [Google Scholar] [CrossRef] [PubMed]
- Vicenzi, E.; Liò, P.; Poli, G. The puzzling role of CXCR4 in human immunodeficiency virus infection. Theranostics 2013, 3, 18. [Google Scholar] [CrossRef]
- Philpott, S.M. HIV-1 coreceptor usage, transmission, and disease progression. Curr. HIV Res. 2003, 1, 217–227. [Google Scholar] [CrossRef]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef] [Green Version]
- Tebit, D.M.; Arts, E.J. Tracking a century of global expansion and evolution of HIV to drive understanding and to combat disease. Lancet Infect. Dis. 2011, 11, 45–56. [Google Scholar] [CrossRef]
- Pollakis, G.; Kang, S.; Kliphuis, A.; Chalaby, M.I.; Goudsmit, J.; Paxton, W.A. N-linked glycosylation of the HIV type-1 gp120 envelope glycoprotein as a major determinant of CCR5 and CXCR4 coreceptor utilization. J. Biol. Chem. 2001, 276, 13433–13441. [Google Scholar] [CrossRef] [Green Version]
- Jo, S.; Song, K.C.; Desaire, H.; MacKerell, A.D., Jr.; Im, W. Glycan Reader: Automated sugar identification and simulation preparation for carbohydrates and glycoproteins. J. Comput. Chem. 2011, 32, 3135–3141. [Google Scholar] [CrossRef] [Green Version]
- Nijmeijer, B.M.; Geijtenbeek, T.B. Negative and positive selection pressure during sexual transmission of transmitted founder HIV-1. Front. Immunol. 2019, 10, 1599. [Google Scholar] [CrossRef] [Green Version]
- Wolf, D.P.; Sokoloski, J.E.; Litt, M. Composition and function of human cervical mucus. Biochim. Et Biophys. Acta (BBA)-Gen. Subj. 1980, 630, 545–558. [Google Scholar] [CrossRef]
- Grivel, J.-C.; Shattock, R.J.; Margolis, L.B. Selective transmission of R5 HIV-1 variants: Where is the gatekeeper? J. Transl. Med. 2011, 9, S6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moulard, M.; Lortat-Jacob, H.; Mondor, I.; Roca, G.; Wyatt, R.; Sodroski, J.; Zhao, L.; Olson, W.; Kwong, P.D.; Sattentau, Q.J. Selective interactions of polyanions with basic surfaces on human immunodeficiency virus type 1 gp120. J. Virol. 2000, 74, 1948–1960. [Google Scholar] [CrossRef] [Green Version]
- Fantini, J.; Hammache, D.; Delézay, O.; Piéroni, G.; Tamalet, C.; Yahi, N. Sulfatide inhibits HIV-1 entry into CD4−/CXCR4+ cells. Virology 1998, 246, 211–220. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Yahi, N.; Tourres, C.; Delezay, O.; Tamalet, C. HIV-1 transmission across the vaginal epithelium. AIDS 1997, 11, 1663–1664. [Google Scholar]
- Berlier, W.; Bourlet, T.; Lawrence, P.; Hamzeh, H.; Lambert, C.; Genin, C.; Verrier, B.; Dieu-Nosjean, M.C.; Pozzetto, B.; Delézay, O. Selective sequestration of X4 isolates by human genital epithelial cells: Implication for virus tropism selection process during sexual transmission of HIV. J. Med. Virol. 2005, 77, 465–474. [Google Scholar] [CrossRef]
- Lawrence, P.; Portran, D.; Terrasse, R.; Palle, S.; Olivier, T.; Fantini, J.; Bourlet, T.; Pozzetto, B.; Delezay, O. Selective transmigration of monocyte-associated HIV-1 across a human cervical monolayer and its modulation by seminal plasma. AIDS 2012, 26, 785–796. [Google Scholar] [CrossRef]
- Margolis, L.; Shattock, R. Selective transmission of CCR5-utilizing HIV-1: The’gatekeeper’problem resolved? Nat. Rev. Microbiol. 2006, 4, 312–317. [Google Scholar] [CrossRef]
- Schutten, M.; Van Baalen, C.; Guillon, C.; Huisman, R.; Boers, P.; Sintnicolaas, K.; Gruters, R.; Osterhaus, A.D. Macrophage tropism of human immunodeficiency virus type 1 facilitates in vivo escape from cytotoxic T-lymphocyte pressure. J. Virol. 2001, 75, 2706–2709. [Google Scholar] [CrossRef] [Green Version]
- Tscherning, C.; Alaeus, A.; Fredriksson, R.; Björndal, Å.; Deng, H.; Littman, D.R.; Fenyö, E.M.; Albert, J. Differences in chemokine coreceptor usage between genetic subtypes of HIV-1. Virology 1998, 241, 181–188. [Google Scholar] [CrossRef]
- Yahi, N.; Fantini, J.; Tourres, C.; Tivoli, N.; Koch, N.; Tamalet, C. Use of drug resistance sequence data for the systematic detection of non-B human immunodeficiency virus type 1 (HIV-1) subtypes: How to create a sentinel site for monitoring the genetic diversity of HIV-1 at a country scale. J. Infect. Dis. 2001, 183, 1311–1317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Righetto, I.; Milani, A.; Cattoli, G.; Filippini, F. Comparative structural analysis of haemagglutinin proteins from type A influenza viruses: Conserved and variable features. BMC Bioinform. 2014, 15, 363. [Google Scholar] [CrossRef] [PubMed]
- Righetto, I.; Filippini, F. Normal modes analysis and surface electrostatics of haemagglutinin proteins as fingerprints for high pathogenic type A influenza viruses. BMC Bioinform. 2020, 21, 354. [Google Scholar] [CrossRef] [PubMed]
- Jimenez-Alberto, A.; Alvarado-Facundo, E.; Ribas-Aparicio, R.M.; Castelán-Vega, J.A. Analysis of adaptation mutants in the hemagglutinin of the influenza A (H1N1) pdm09 virus. PLoS ONE 2013, 8, e70005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weis, W.; Brown, J.; Cusack, S.; Paulson, J.; Skehel, J.; Wiley, D. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 1988, 333, 426–431. [Google Scholar] [CrossRef]
- Kumlin, U.; Olofsson, S.; Dimock, K.; Arnberg, N. Sialic acid tissue distribution and influenza virus tropism. Influenza Other Respir. Viruses 2008, 2, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Franca, M.; Stallknecht, D.; Howerth, E. Expression and distribution of sialic acid influenza virus receptors in wild birds. Avian Pathol. 2013, 42, 60–71. [Google Scholar] [CrossRef]
- Qi, L.; Kash, J.C.; Dugan, V.G.; Wang, R.; Jin, G.; Cunningham, R.E.; Taubenberger, J.K. Role of sialic acid binding specificity of the 1918 influenza virus hemagglutinin protein in virulence and pathogenesis for mice. J. Virol. 2009, 83, 3754–3761. [Google Scholar] [CrossRef] [Green Version]
- Ayora-Talavera, G. Sialic acid receptors: Focus on their role in influenza infection. J. Recept. Ligand Channel Res. 2018, 10, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Xiong, X.; Coombs, P.J.; Martin, S.R.; Liu, J.; Xiao, H.; McCauley, J.W.; Locher, K.; Walker, P.A.; Collins, P.J.; Kawaoka, Y.; et al. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 2013, 497, 392–396. [Google Scholar] [CrossRef]
- Zhu, X.; Yu, W.; McBride, R.; Li, Y.; Chen, L.M.; Donis, R.O.; Tong, S.; Paulson, J.C.; Wilson, I.A. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc. Natl. Acad. Sci. USA 2013, 110, 1458–1463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinstein, R.A. Planning for epidemics--the lessons of SARS. N. Engl. J. Med. 2004, 350, 2332–2334. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Zhou, P.; Yang, X.L.; Wang, X.G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.R.; Zhu, Y.; Li, B.; Huang, C.L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.; Liu, D.X.; Tam, J.P. Lipid rafts are involved in SARS-CoV entry into Vero E6 cells. Biochem. Biophys. Res. Commun. 2008, 369, 344–349. [Google Scholar] [CrossRef]
- Li, G.-M.; Li, Y.-G.; Yamate, M.; Li, S.-M.; Ikuta, K. Lipid rafts play an important role in the early stage of severe acute respiratory syndrome-coronavirus life cycle. Microbes Infect. 2007, 9, 96–102. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Hulswit, R.J.; Widjaja, I.; Raj, V.S.; McBride, R.; Peng, W.; Widagdo, W.; Tortorici, M.A.; Van Dieren, B.; Lang, Y. Identification of sialic acid-binding function for the Middle East respiratory syndrome coronavirus spike glycoprotein. Proc. Natl. Acad. Sci. USA 2017, 114, E8508–E8517. [Google Scholar] [CrossRef] [Green Version]
- Sun, X.-L. The role of cell surface sialic acids for SARS-CoV-2 infection. Glycobiology 2021, 31, 1245–1253. [Google Scholar] [CrossRef]
- Pirone, L.; Del Gatto, A.; Di Gaetano, S.; Saviano, M.; Capasso, D.; Zaccaro, L.; Pedone, E. A multi-targeting approach to fight SARS-CoV-2 attachment. Front. Mol. Biosci. 2020, 7, 186. [Google Scholar] [CrossRef]
- Raj, V.S.; Mou, H.; Smits, S.L.; Dekkers, D.H.; Müller, M.A.; Dijkman, R.; Muth, D.; Demmers, J.A.; Zaki, A.; Fouchier, R.A. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 2013, 495, 251–254. [Google Scholar] [CrossRef] [Green Version]
- Hatmal, M.M.M.; Alshaer, W.; Al-Hatamleh, M.A.; Hatmal, M.; Smadi, O.; Taha, M.O.; Oweida, A.J.; Boer, J.C.; Mohamud, R.; Plebanski, M. Comprehensive structural and molecular comparison of spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and their interactions with ACE2. Cells 2020, 9, 2638. [Google Scholar] [CrossRef] [PubMed]
- Boschi, C.; Scheim, D.E.; Bancod, A.; Militello, M.; Bideau, M.L.; Colson, P.; Fantini, J.; Scola, B.L. SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects. Int. J. Mol. Sci. 2022, 23, 15480. [Google Scholar] [CrossRef] [PubMed]
- Stencel-Baerenwald, J.E.; Reiss, K.; Reiter, D.M.; Stehle, T.; Dermody, T.S. The sweet spot: Defining virus–sialic acid interactions. Nat. Rev. Microbiol. 2014, 12, 739–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guérin, P.; Yahi, N.; Azzaz, F.; Chahinian, H.; Sabatier, J.M.; Fantini, J. Structural Dynamics of the SARS-CoV-2 Spike Protein: A 2-Year Retrospective Analysis of SARS-CoV-2 Variants (from Alpha to Omicron) Reveals an Early Divergence between Conserved and Variable Epitopes. Molecules 2022, 27, 3851. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef] [PubMed]
- Lazarevic, I.; Pravica, V.; Miljanovic, D.; Cupic, M. Immune evasion of SARS-CoV-2 emerging variants: What have we learnt so far? Viruses 2021, 13, 1192. [Google Scholar] [CrossRef]
- Hu, J.; Peng, P.; Cao, X.; Wu, K.; Chen, J.; Wang, K.; Tang, N.; Huang, A.-l. Increased immune escape of the new SARS-CoV-2 variant of concern Omicron. Cell. Mol. Immunol. 2022, 19, 293–295. [Google Scholar] [CrossRef]
- Barton, M.I.; MacGowan, S.A.; Kutuzov, M.A.; Dushek, O.; Barton, G.J.; Van Der Merwe, P.A. Effects of common mutations in the SARS-CoV-2 Spike RBD and its ligand, the human ACE2 receptor on binding affinity and kinetics. elife 2021, 10, e70658. [Google Scholar] [CrossRef]
- Moulana, A.; Dupic, T.; Phillips, A.M.; Chang, J.; Nieves, S.; Roffler, A.A.; Greaney, A.J.; Starr, T.N.; Bloom, J.D.; Desai, M.M. Compensatory epistasis maintains ACE2 affinity in SARS-CoV-2 Omicron BA.1. Nat. Commun. 2022, 13, 7011. [Google Scholar] [CrossRef]
- Fantini, J.; Yahi, N.; Colson, P.; Chahinian, H.; La Scola, B.; Raoult, D. The puzzling mutational landscape of the SARS-2-variant Omicron. J. Med. Virol. 2022, 94, 2019–2025. [Google Scholar] [CrossRef]
- Pascarella, S.; Ciccozzi, M.; Bianchi, M.; Benvenuto, D.; Cauda, R.; Cassone, A. The electrostatic potential of the Omicron variant spike is higher than in Delta and Delta-plus variants: A hint to higher transmissibility? J. Med. Virol. 2021, 94, 1277–1280. [Google Scholar] [CrossRef] [PubMed]
- Mykytyn, A.Z.; Rissmann, M.; Kok, A.; Rosu, M.E.; Schipper, D.; Breugem, T.I.; van den Doel, P.B.; Chandler, F.; Bestebroer, T.; de Wit, M. Antigenic cartography of SARS-CoV-2 reveals that Omicron BA.1 and BA.2 are antigenically distinct. Sci. Immunol. 2022, 7, eabq4450. [Google Scholar] [PubMed]
- Smith, D.J.; Lapedes, A.S.; De Jong, J.C.; Bestebroer, T.M.; Rimmelzwaan, G.F.; Osterhaus, A.D.; Fouchier, R.A. Mapping the antigenic and genetic evolution of influenza virus. Science 2004, 305, 371–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, M.; Zhao, X.; Hua, S.; Du, X.; Peng, Y.; Li, X.; Lan, Y.; Wang, D.; Wu, A.; Shu, Y. Antigenic patterns and evolution of the human influenza A (H1N1) virus. Sci. Rep. 2015, 5, 14171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCallum, M.; De Marco, A.; Lempp, F.A.; Tortorici, M.A.; Pinto, D.; Walls, A.C.; Beltramello, M.; Chen, A.; Liu, Z.; Zatta, F. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell 2021, 184, 2332–2347.e16. [Google Scholar] [CrossRef]
- Focosi, D.; Novazzi, F.; Baj, A.; Maggi, F. Monkeypox: An international epidemic. Rev. Med. Virol. 2022, 32, e2392. [Google Scholar] [CrossRef]
- Das, T.; Mukhopadhyay, C. Identification of possible binding modes of SARS-CoV-2 spike N-terminal domain for ganglioside GM1. Chem. Phys. Lett. 2022, 812, 140260. [Google Scholar] [CrossRef]
- Liu, L.; Wang, P.; Nair, M.S.; Yu, J.; Rapp, M.; Wang, Q.; Luo, Y.; Chan, J.F.W.; Sahi, V.; Figueroa, A.; et al. Potent neutralizing antibodies against multiple epitopes on SARS-CoV-2 spike. Nature 2020, 584, 450–456. [Google Scholar] [CrossRef]
- Azzaz, F.; Yahi, N.; Chahinian, H.; Fantini, J. The Epigenetic Dimension of Protein Structure Is an Intrinsic Weakness of the AlphaFold Program. Biomolecules 2022, 12, 1527. [Google Scholar] [CrossRef]
- Shantier, S.W.; Mustafa, M.I.; Abdelmoneim, A.H.; Fadl, H.A.; Elbager, S.G.; Makhawi, A.M. Novel multi epitope-based vaccine against monkeypox virus: Vaccinomic approach. Sci. Rep. 2022, 12, 15983. [Google Scholar] [CrossRef]
- Baba, M.; Snoeck, R.; Pauwels, R.; De Clercq, E. Sulfated polysaccharides are potent and selective inhibitors of various enveloped viruses, including herpes simplex virus, cytomegalovirus, vesicular stomatitis virus, and human immunodeficiency virus. Antimicrob. Agents Chemother. 1988, 32, 1742–1745. [Google Scholar] [CrossRef] [Green Version]
- Lin, L.-T.; Chen, T.-Y.; Lin, S.-C.; Chung, C.-Y.; Lin, T.-C.; Wang, G.-H.; Anderson, R.; Lin, C.-C.; Richardson, C.D. Broad-spectrum antiviral activity of chebulagic acid and punicalagin against viruses that use glycosaminoglycans for entry. BMC Microbiol. 2013, 13, 187. [Google Scholar] [CrossRef] [PubMed]
- Yahi, N.; Sabatier, J.M.; Nickel, P.; Mabrouk, K.; Gonzalez-Scarano, F.; Fantini, J. Suramin inhibits binding of the V3 region of HIV-1 envelope glycoprotein gp120 to galactosylceramide, the receptor for HIV-1 gp120 on human colon epithelial cells. J. Biol. Chem. 1994, 269, 24349–24353. [Google Scholar] [CrossRef] [PubMed]
- Delézay, O.; Hammache, D.; Fantini, J.; Yahi, N. SPC3, a V3 loop-derived synthetic peptide inhibitor of HIV-1 infection, binds to cell surface glycosphingolipids. Biochemistry 1996, 35, 15663–15671. [Google Scholar] [CrossRef]
- Yahi, N.; Fantini, J.; Baghdiguian, S.; Mabrouk, K.; Tamalet, C.; Rochat, H.; Van Rietschoten, J.; Sabatier, J.-M. SPC3, a synthetic peptide derived from the V3 domain of human immunodeficiency virus type 1 (HIV-1) gp120, inhibits HIV-1 entry into CD4+ and CD4-cells by two distinct mechanisms. Proc. Natl. Acad. Sci. USA 1995, 92, 4867–4871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yahi, N.; Sabatier, J.M.; Baghdiguian, S.; Gonzalez-Scarano, F.; Fantini, J. Synthetic multimeric peptides derived from the principal neutralization domain (V3 loop) of human immunodeficiency virus type 1 (HIV-1) gp120 bind to galactosylceramide and block HIV-1 infection in a human CD4-negative mucosal epithelial cell line. J. Virol. 1995, 69, 320–325. [Google Scholar] [CrossRef] [Green Version]
- Faroux-Corlay, B.; Greiner, J.; Terreux, R.; Cabrol-Bass, D.; Aubertin, A.-M.; Vierling, P.; Fantini, J. Amphiphilic anionic analogues of galactosylceramide: Synthesis, anti-HIV-1 activity, and gp120 binding. J. Med. Chem. 2001, 44, 2188–2203. [Google Scholar] [CrossRef]
- Faroux-Corlay, B.; Clary, L.; Gadras, C.; Hammache, D.; Greiner, J.; Santaella, C.; Aubertin, A.-M.; Vierling, P.; Fantini, J. Synthesis of single-and double-chain fluorocarbon and hydrocarbon galactosyl amphiphiles and their anti-HIV-1 activity. Carbohydr. Res. 2000, 327, 223–260. [Google Scholar] [CrossRef]
- Kensinger, R.D.; Yowler, B.C.; Benesi, A.J.; Schengrund, C.-L. Synthesis of novel, multivalent glycodendrimers as ligands for HIV-1 gp120. Bioconjugate Chem. 2004, 15, 349–358. [Google Scholar] [CrossRef]
- Garg, H.; Francella, N.; Tony, K.A.; Augustine, L.A.; Barchi Jr, J.J.; Fantini, J.; Puri, A.; Mootoo, D.R.; Blumenthal, R. Glycoside analogs of β-galactosylceramide, a novel class of small molecule antiviral agents that inhibit HIV-1 entry. Antivir. Res. 2008, 80, 54–61. [Google Scholar] [CrossRef] [Green Version]
- Fantini, J.; Hammache, D.; Delézay, O.; Yahi, N.; André-Barrès, C.; Rico-Lattes, I.; Lattes, A. Synthetic soluble analogs of galactosylceramide (GalCer) bind to the V3 domain of HIV-1 gp120 and inhibit HIV-1-induced fusion and entry. J. Biol. Chem. 1997, 272, 7245–7252. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautret, P.; Lagier, J.-C.; Parola, P.; Meddeb, L.; Mailhe, M.; Doudier, B.; Courjon, J.; Giordanengo, V.; Vieira, V.E.; Dupont, H.T. Hydroxychloroquine and azithromycin as a treatment of COVID-19: Results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents 2020, 56, 105949. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Di Scala, C.; Chahinian, H.; Yahi, N. Structural and molecular modelling studies reveal a new mechanism of action of chloroquine and hydroxychloroquine against SARS-CoV-2 infection. Int. J. Antimicrob. Agents 2020, 55, 105960. [Google Scholar] [CrossRef] [PubMed]
- Fantini, J.; Chahinian, H.; Yahi, N. Synergistic antiviral effect of hydroxychloroquine and azithromycin in combination against SARS-CoV-2: What molecular dynamics studies of virus-host interactions reveal. Int. J. Antimicrob. Agents 2020, 56, 106020. [Google Scholar] [CrossRef]
- Andreani, J.; Le Bideau, M.; Duflot, I.; Jardot, P.; Rolland, C.; Boxberger, M.; Wurtz, N.; Rolain, J.-M.; Colson, P.; La Scola, B. In vitro testing of combined hydroxychloroquine and azithromycin on SARS-CoV-2 shows synergistic effect. Microb. Pathog. 2020, 145, 104228. [Google Scholar] [CrossRef]
- Jans, D.A.; Wagstaff, K.M. Ivermectin as a broad-spectrum host-directed antiviral: The real deal? Cells 2020, 9, 2100. [Google Scholar] [CrossRef]
- Colson, P.; Raoult, D. Fighting viruses with antibiotics: An overlooked path. Int. J. Antimicrob. Agents 2016, 48, 349. [Google Scholar] [CrossRef]
- Khoshnood, S.; Shirani, M.; Dalir, A.; Moradi, M.; Haddadi, M.H.; Sadeghifard, N.; Birjandi, F.S.; Yashmi, I.; Heidary, M. Antiviral effects of azithromycin: A narrative review. Biomed. Pharmacother. 2022, 147, 112682. [Google Scholar] [CrossRef]
- Rizzo, E. Ivermectin, antiviral properties and COVID-19: A possible new mechanism of action. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2020, 393, 1153–1156. [Google Scholar] [CrossRef]
- Rodrigo, C.; Fernando, S.D.; Rajapakse, S. Clinical evidence for repurposing chloroquine and hydroxychloroquine as antiviral agents: A systematic review. Clin. Microbiol. Infect. 2020, 26, 979–987. [Google Scholar] [CrossRef]
- Tan, C.W.; Sam, I.-C.; Chong, W.L.; Lee, V.S.; Chan, Y.F. Polysulfonate suramin inhibits Zika virus infection. Antivir. Res. 2017, 143, 186–194. [Google Scholar] [CrossRef]
- Gustafson, K.R.; Cardellina, J.H.; Fuller, R.W.; Weislow, O.S.; Kiser, R.F.; Snader, K.M.; Patterson, G.M.; Boyd, M.R. AIDS-antiviral sulfolipids from cyanobacteria (blue-green algae). JNCI J. Natl. Cancer Inst. 1989, 81, 1254–1258. [Google Scholar] [CrossRef]
- Xu, M.; Pradhan, M.; Gorshkov, K.; Petersen, J.D.; Shen, M.; Guo, H.; Zhu, W.; Klumpp-Thomas, C.; Michael, S.; Itkin, M. A high throughput screening assay for inhibitors of SARS-CoV-2 pseudotyped particle entry. Slas Discov. 2022, 27, 86–94. [Google Scholar] [CrossRef]
- Steffen, I.; Simmons, G. Pseudotyping viral vectors with emerging virus envelope proteins. Curr. Gene Ther. 2016, 16, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Carter-Timofte, M.E.; Arulanandam, R.; Kurmasheva, N.; Fu, K.; Laroche, G.; Taha, Z.; van Der Horst, D.; Cassin, L.; van der Sluis, R.M.; Palermo, E. Antiviral potential of the antimicrobial drug atovaquone against SARS-CoV-2 and emerging variants of concern. ACS Infect. Dis. 2021, 7, 3034–3051. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Sun, J.; Wang, K.; Wang, H.; Wu, S.; Bao, L.; He, W.; Wang, D.; Zhu, A.; Zhang, T. Repurposing carrimycin as an antiviral agent against human coronaviruses, including the currently pandemic SARS-CoV-2. Acta Pharm. Sin. B 2021, 11, 2850–2858. [Google Scholar] [CrossRef] [PubMed]
- Du, X.; Zuo, X.; Meng, F.; Han, C.; Ouyang, W.; Han, Y.; Gu, Y.; Zhao, X.; Xu, F.; Qin, F.X. Direct inhibitory effect on viral entry of influenza A and SARS-CoV-2 viruses by azithromycin. Cell Prolif. 2021, 54, e12953. [Google Scholar] [CrossRef] [PubMed]
- Ou, T.; Mou, H.; Zhang, L.; Ojha, A.; Choe, H.; Farzan, M. Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog. 2021, 17, e1009212. [Google Scholar] [CrossRef]
- Henß, L.; Beck, S.; Weidner, T.; Biedenkopf, N.; Sliva, K.; Weber, C.; Becker, S.; Schnierle, B.S. Suramin is a potent inhibitor of Chikungunya and Ebola virus cell entry. Virol. J. 2016, 13, 149. [Google Scholar] [CrossRef] [Green Version]
- Rojo, J.; Delgado, R. Glycodendritic structures: Promising new antiviral drugs. J. Antimicrob. Chemother. 2004, 54, 579–581. [Google Scholar] [CrossRef] [Green Version]
- Niaee, M.S.; Namdar, P.; Allami, A.; Zolghadr, L.; Javadi, A.; Karampour, A.; Varnaseri, M.; Bijani, B.; Cheraghi, F.; Naderi, Y. Ivermectin as an adjunct treatment for hospitalized adult COVID-19 patients: A randomized multi-center clinical trial. Asian Pac. J. Trop. Med. 2021, 14, 266. [Google Scholar]
- Mahmud, R.; Rahman, M.M.; Alam, I.; Ahmed, K.G.U.; Kabir, A.H.; Sayeed, S.J.B.; Rassel, M.A.; Monayem, F.B.; Islam, M.S.; Islam, M.M. Ivermectin in combination with doxycycline for treating COVID-19 symptoms: A randomized trial. J. Int. Med. Res. 2021, 49, 03000605211013550. [Google Scholar] [CrossRef] [PubMed]
- Seet, R.C.S.; Quek, A.M.L.; Ooi, D.S.Q.; Sengupta, S.; Lakshminarasappa, S.R.; Koo, C.Y.; So, J.B.Y.; Goh, B.C.; Loh, K.S.; Fisher, D.; et al. Positive impact of oral hydroxychloroquine and povidone-iodine throat spray for COVID-19 prophylaxis: An open-label randomized trial. Int. J. Infect. Dis. 2021, 106, 314–322. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Y.; Agostinis, P.; Rabson, A.; Melino, G.; Carafoli, E.; Shi, Y.; Sun, E. Is hydroxychloroquine beneficial for COVID-19 patients? Cell Death Dis. 2020, 11, 512. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fantini, J.; Azzaz, F.; Chahinian, H.; Yahi, N. Electrostatic Surface Potential as a Key Parameter in Virus Transmission and Evolution: How to Manage Future Virus Pandemics in the Post-COVID-19 Era. Viruses 2023, 15, 284. https://doi.org/10.3390/v15020284
Fantini J, Azzaz F, Chahinian H, Yahi N. Electrostatic Surface Potential as a Key Parameter in Virus Transmission and Evolution: How to Manage Future Virus Pandemics in the Post-COVID-19 Era. Viruses. 2023; 15(2):284. https://doi.org/10.3390/v15020284
Chicago/Turabian StyleFantini, Jacques, Fodil Azzaz, Henri Chahinian, and Nouara Yahi. 2023. "Electrostatic Surface Potential as a Key Parameter in Virus Transmission and Evolution: How to Manage Future Virus Pandemics in the Post-COVID-19 Era" Viruses 15, no. 2: 284. https://doi.org/10.3390/v15020284




