Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Purification
2.2. Cryo-EM Imaging
2.3. Image Processing of the P22 Particles
2.4. Atomic Model Building and Refinement
3. Results
3.1. Cryo-EM Reconstruction of P22 Particles in the Two Bands of the Gradient
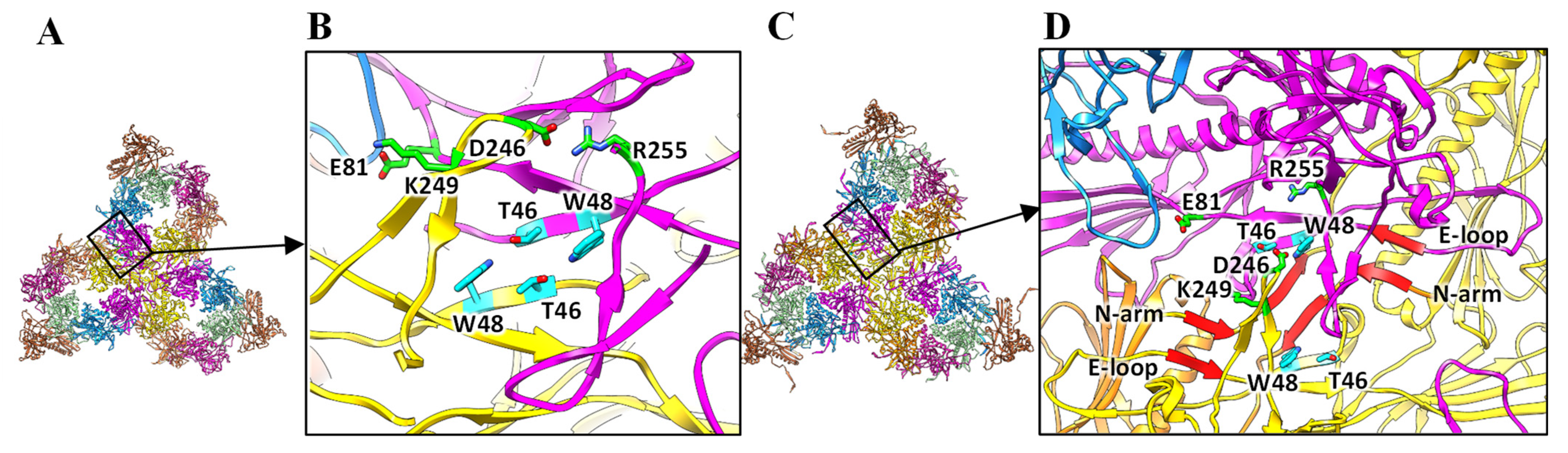
3.2. Structure of the Coat Protein and Assembly of the Procapsid
3.3. Interactions between the Scaffolding Protein and the Inner Procapsid
3.4. Conformational Changes in the Coat Protein during Viral Maturation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Conway, J.F.; Wikoff, W.R.; Cheng, N.; Duda, R.L.; Hendrix, R.W.; Johnson, J.E.; Steven, A.C. Virus maturation involving large subunit rotations and local refolding. Science 2001, 292, 744–748. [Google Scholar] [CrossRef]
- Suhanovsky, M.M.; Teschke, C.M. Nature’s favorite building block: Deciphering folding and capsid assembly of proteins with the HK97-fold. Virology 2015, 479, 487–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, D.H.; Baker, M.L.; Hryc, C.F.; DiMaio, F.; Jakana, J.; Wu, W.; Dougherty, M.; Haase-Pettingell, C.; Schmid, M.F.; Jiang, W.; et al. Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. USA 2011, 108, 1355–1360. [Google Scholar] [CrossRef] [Green Version]
- Michaud, J.M.; Thompson, L.R.; Kaul, D.; Espinoza, J.L.; Richter, R.A.; Xu, Z.Z.; Lee, C.; Pham, K.M.; Beall, C.M.; Malfatti, F.; et al. Taxon-specific aerosolization of bacteria and viruses in an experimental ocean-atmosphere mesocosm. Nat. Commun. 2018, 9, 2017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendrix, R.W.; Smith, M.C.; Burns, R.N.; Ford, M.E.; Hatfull, G.F. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc. Natl. Acad. Sci. USA 1999, 96, 2192–2197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmond, G.P.; Fineran, P.C. A century of the phage: Past, present and future. Nat Rev. Microbiol. 2015, 13, 777–786. [Google Scholar] [CrossRef]
- Cardone, G.; Heymann, J.B.; Cheng, N.; Trus, B.L.; Steven, A.C. Procapsid assembly, maturation, nuclear exit: Dynamic steps in the production of infectious herpesvirions. Adv. Exp. Med. Biol. 2012, 726, 423–439. [Google Scholar]
- Prevelige, P.E.; Fane, B.A. Building the machines: Scaffolding protein functions during bacteriophage morphogenesis. Adv. Exp. Med. Biol. 2012, 726, 325–350. [Google Scholar]
- Guo, F.; Liu, Z.; Fang, P.A.; Zhang, Q.; Wright, E.T.; Wu, W.; Zhang, C.; Vago, F.; Ren, Y.; Jakana, J.; et al. Capsid expansion mechanism of bacteriophage T7 revealed by multistate atomic models derived from cryo-EM reconstructions. Proc. Natl. Acad. Sci. USA 2014, 111, E4606-14. [Google Scholar] [CrossRef] [Green Version]
- Ignatiou, A.; Brasiles, S.; El Sadek Fadel, M.; Burger, J.; Mielke, T.; Topf, M.; Tavares, P.; Orlova, E.V. Structural transitions during the scaffolding-driven assembly of a viral capsid. Nat. Commun. 2019, 10, 4840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bayfield, O.W.; Klimuk, E.; Winkler, D.C.; Hesketh, E.L.; Chechik, M.; Cheng, N.; Dykeman, E.C.; Minakhin, L.; Ranson, N.A.; Severinov, K.; et al. Cryo-EM structure and in vitro DNA packaging of a thermophilic virus with supersized T=7 capsids. Proc. Natl. Acad. Sci. USA 2019, 116, 3556–3561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gertsman, I.; Gan, L.; Guttman, M.; Lee, K.; Speir, J.A.; Duda, R.L.; Hendrix, R.W.; Komives, E.A.; Johnson, J.E. An unexpected twist in viral capsid maturation. Nature 2009, 458, 646–650. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zeng, J.; Wang, J. Structural basis of bacteriophage lambda capsid maturation. Structure 2022, 30, 637–645.e3. [Google Scholar] [CrossRef] [PubMed]
- Duda, R.L.; Teschke, C.M. The amazing HK97 fold: Versatile results of modest differences. Curr. Opin. Virol. 2019, 36, 9–16. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Liljas, L.; Duda, R.L.; Tsuruta, H.; Hendrix, R.W.; Johnson, J.E. Topologically linked protein rings in the bacteriophage HK97 capsid. Science 2000, 289, 2129–2133. [Google Scholar] [CrossRef] [Green Version]
- Parent, K.N.; Khayat, R.; Tu, L.H.; Suhanovsky, M.M.; Cortines, J.R.; Teschke, C.M.; Johnson, J.E.; Baker, T.S. P22 coat protein structures reveal a novel mechanism for capsid maturation: Stability without auxiliary proteins or chemical crosslinks. Structure 2010, 18, 390–401. [Google Scholar] [CrossRef] [Green Version]
- Casjens, S.R.; Molineux, I.J. Short noncontractile tail machines: Adsorption and DNA delivery by podoviruses. Adv. Exp. Med. Biol. 2012, 726, 143–179. [Google Scholar]
- O’Neil, A.; Reichhardt, C.; Johnson, B.; Prevelige, P.E.; Douglas, T. Genetically programmed in vivo packaging of protein cargo and its controlled release from bacteriophage P22. Angew. Chem. Int. Ed. Engl. 2011, 50, 7425–7428. [Google Scholar] [CrossRef]
- Patterson, D.P.; Schwarz, B.; Waters, R.S.; Gedeon, T.; Douglas, T. Encapsulation of an enzyme cascade within the bacteriophage P22 virus-like particle. ACS Chem. Biol. 2014, 9, 359–365. [Google Scholar] [CrossRef]
- McCoy, K.; Selivanovitch, E.; Luque, D.; Lee, B.; Edwards, E.; Caston, J.R.; Douglas, T. Cargo Retention inside P22 Virus-Like Particles. Biomacromolecules 2018, 19, 3738–3746. [Google Scholar] [CrossRef]
- King, J.; Casjens, S. Catalytic head assembling protein in virus morphogenesis. Nature 1974, 251, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Parker, M.H.; Weigele, P.; Casjens, S.; Prevelige, P.E., Jr.; Krishna, N.R. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J. Mol. Biol. 2000, 297, 1195–1202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hryc, C.F.; Chen, D.H.; Afonine, P.V.; Jakana, J.; Wang, Z.; Haase-Pettingell, C.; Jiang, W.; Adams, P.D.; King, J.A.; Schmid, M.F.; et al. Accurate model annotation of a near-atomic resolution cryo-EM map. Proc. Natl. Acad. Sci. USA 2017, 114, 3103–3108. [Google Scholar] [CrossRef] [PubMed]
- Prasad, B.V.; Prevelige, P.E.; Marietta, E.; Chen, R.O.; Thomas, D.; King, J.; Chiu, W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J. Mol. Biol. 1993, 231, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Thuman-Commike, P.A.; Greene, B.; Jakana, J.; Prasad, B.V.; King, J.; Prevelige, P.E., Jr.; Chiu, W. Three-dimensional structure of scaffolding-containing phage p22 procapsids by electron cryo-microscopy. J. Mol. Biol. 1996, 260, 85–98. [Google Scholar] [CrossRef]
- Jiang, W.; Li, Z.; Zhang, Z.; Baker, M.L.; Prevelige, P.E., Jr.; Chiu, W. Coat protein fold and maturation transition of bacteriophage P22 seen at subnanometer resolutions. Nat. Struct. Biol. 2003, 10, 131–135. [Google Scholar] [CrossRef]
- Rizzo, A.A.; Suhanovsky, M.M.; Baker, M.L.; Fraser, L.C.; Jones, L.M.; Rempel, D.L.; Gross, M.L.; Chiu, W.; Alexandrescu, A.T.; Teschke, C.M. Multiple functional roles of the accessory I-domain of bacteriophage P22 coat protein revealed by NMR structure and CryoEM modeling. Structure 2014, 22, 830–841. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhou, N.; Chen, W.; Zhu, B.; Wang, X.; Xu, B.; Wang, J.; Liu, H.; Cheng, L. Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV. J. Mol. Biol. 2017, 429, 79–87. [Google Scholar] [CrossRef]
- Li, X.; Liu, H.; Cheng, L. Symmetry-mismatch reconstruction of genomes and associated proteins within icosahedral viruses using cryo-EM. Biophys Rep. 2016, 2, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Thuman-Commike, P.A.; Chiu, W. Improved common line-based icosahedral particle image orientation estimation algorithms. Ultramicroscopy 1997, 68, 231–255. [Google Scholar] [CrossRef]
- Kivioja, T.; Ravantti, J.; Verkhovsky, A.; Ukkonen, E.; Bamford, D. Local average intensity-based method for identifying spherical particles in electron micrographs. J. Struct. Biol. 2000, 131, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Cheng, L.; Zeng, S.; Cai, C.; Zhou, Z.H.; Yang, Q. Symmetry-adapted spherical harmonics method for high-resolution 3D single-particle reconstructions. J. Struct. Biol. 2008, 161, 64–73. [Google Scholar] [CrossRef]
- DiMaio, F.; Tyka, M.D.; Baker, M.L.; Chiu, W.; Baker, D. Refinement of protein structures into low-resolution density maps using rosetta. J. Mol. Biol. 2009, 392, 181–190. [Google Scholar] [CrossRef]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Cryst. D. Biol. Cryst. 2010, 66, 486–501. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D Biol. Cryst. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein. Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef] [Green Version]
- de Sousa, P.C., Jr.; Tuma, R.; Prevelige, P.E., Jr.; Silva, J.L.; Foguel, D. Cavity defects in the procapsid of bacteriophage P22 and the mechanism of capsid maturation. J. Mol. Biol. 1999, 287, 527–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gordon, C.L.; King, J. Temperature-sensitive mutations in the phage P22 coat protein which interfere with polypeptide chain folding. J. Biol. Chem. 1993, 268, 9358–9368. [Google Scholar] [CrossRef] [PubMed]
- Parker, M.H.; Casjens, S.; Prevelige, P.E., Jr. Functional domains of bacteriophage P22 scaffolding protein. J. Mol. Biol. 1998, 281, 69–79. [Google Scholar] [CrossRef]
- Cortines, J.R.; Motwani, T.; Vyas, A.A.; Teschke, C.M. Highly specific salt bridges govern bacteriophage P22 icosahedral capsid assembly: Identification of the site in coat protein responsible for interaction with scaffolding protein. J. Virol. 2014, 88, 5287–5297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cortines, J.R.; Weigele, P.R.; Gilcrease, E.B.; Casjens, S.R.; Teschke, C.M. Decoding bacteriophage P22 assembly: Identification of two charged residues in scaffolding protein responsible for coat protein interaction. Virology. 2011, 421, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigele, P.R.; Sampson, L.; Winn-Stapley, D.; Casjens, S.R. Molecular genetics of bacteriophage P22 scaffolding protein’s functional domains. J. Mol. Biol. 2005, 348, 831–844. [Google Scholar] [CrossRef] [PubMed]
- Lander, G.C.; Tang, L.; Casjens, S.R.; Gilcrease, E.B.; Prevelige, P.; Poliakov, A.; Potter, C.S.; Carragher, B.; Johnson, J.E. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science 2006, 312, 1791–1795. [Google Scholar] [CrossRef] [Green Version]
- Earnshaw, W.; King, J. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J. Mol. Biol. 1978, 126, 721–747. [Google Scholar] [CrossRef]
- Huet, A.; Duda, R.L.; Boulanger, P.; Conway, J.F. Capsid expansion of bacteriophage T5 revealed by high resolution cryoelectron microscopy. Proc. Natl. Acad. Sci. USA 2019, 116, 21037–21046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, Q.; Tang, W.C.; Fokine, A.; Mahalingam, M.; Shao, Q.; Rossmann, M.G.; Rao, V.B. Structures of a large prolate virus capsid in unexpanded and expanded states generate insights into the icosahedral virus assembly. Proc. Natl. Acad. Sci. USA 2022, 119, e2203272119. [Google Scholar] [CrossRef]
- Helgstrand, C.; Wikoff, W.R.; Duda, R.L.; Hendrix, R.W.; Johnson, J.E.; Liljas, L. The refined structure of a protein catenane: The HK97 bacteriophage capsid at 3.44 A resolution. J. Mol. Biol. 2003, 334, 885–899. [Google Scholar] [CrossRef] [PubMed]
- Black, L.W.; Rao, V.B. Structure, assembly, and DNA packaging of the bacteriophage T4 head. Adv. Virus Res. 2012, 82, 119–153. [Google Scholar]
- Medina, E.; Wieczorek, D.; Medina, E.M.; Yang, Q.; Feiss, M.; Catalano, C.E. Assembly and maturation of the bacteriophage lambda procapsid: gpC is the viral protease. J. Mol. Biol. 2010, 401, 813–830. [Google Scholar] [CrossRef]
- Fokine, A.; Rossmann, M.G. Common Evolutionary Origin of Procapsid Proteases, Phage Tail Tubes, and Tubes of Bacterial Type VI Secretion Systems. Structure 2016, 24, 1928–1935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preux, O.; Durand, D.; Huet, A.; Conway, J.F.; Bertin, A.; Boulogne, C.; Drouin-Wahbi, J.; Trevarin, D.; Perez, J.; Vachette, P.; et al. A two-state cooperative expansion converts the procapsid shell of bacteriophage T5 into a highly stable capsid isomorphous to the final virion head. J. Mol. Biol. 2013, 425, 1999–2014. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, R.W.; Johnson, J.E. Bacteriophage HK97 capsid assembly and maturation. Adv. Exp. Med. Biol. 2012, 726, 351–363. [Google Scholar] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, H.; Zhou, J.; Yang, F.; Liu, Z.; Song, J.; Chen, W.; Liu, H.; Cheng, L. Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures. Viruses 2023, 15, 355. https://doi.org/10.3390/v15020355
Xiao H, Zhou J, Yang F, Liu Z, Song J, Chen W, Liu H, Cheng L. Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures. Viruses. 2023; 15(2):355. https://doi.org/10.3390/v15020355
Chicago/Turabian StyleXiao, Hao, Junquan Zhou, Fan Yang, Zheng Liu, Jingdong Song, Wenyuan Chen, Hongrong Liu, and Lingpeng Cheng. 2023. "Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures" Viruses 15, no. 2: 355. https://doi.org/10.3390/v15020355



