Epitope Coverage of Anti-SARS-CoV-2 Nucleocapsid IgA and IgG Antibodies Correlates with Protection against Re-Infection by New Variants in Subsequent Waves of the COVID-19 Pandemic
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cohort Information and Sample Collection
2.2. Protein Microarrays
2.3. Serological Assays
2.4. Bioinformatic Analysis
2.4.1. Image Analysis: Raw Data Extraction and Data Pre-Processing
2.4.2. Statistical Tests
3. Results
3.1. Seroprevelance
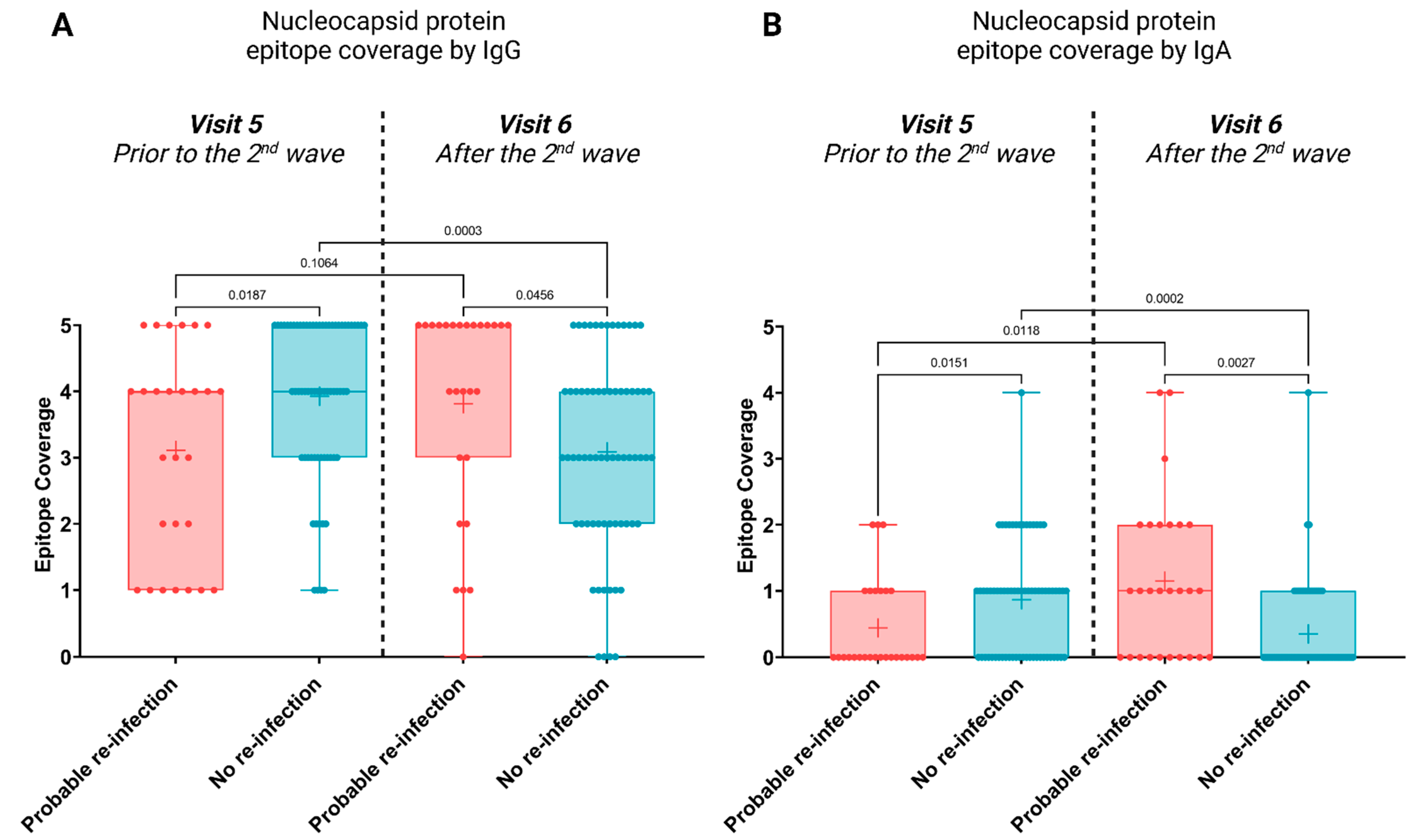
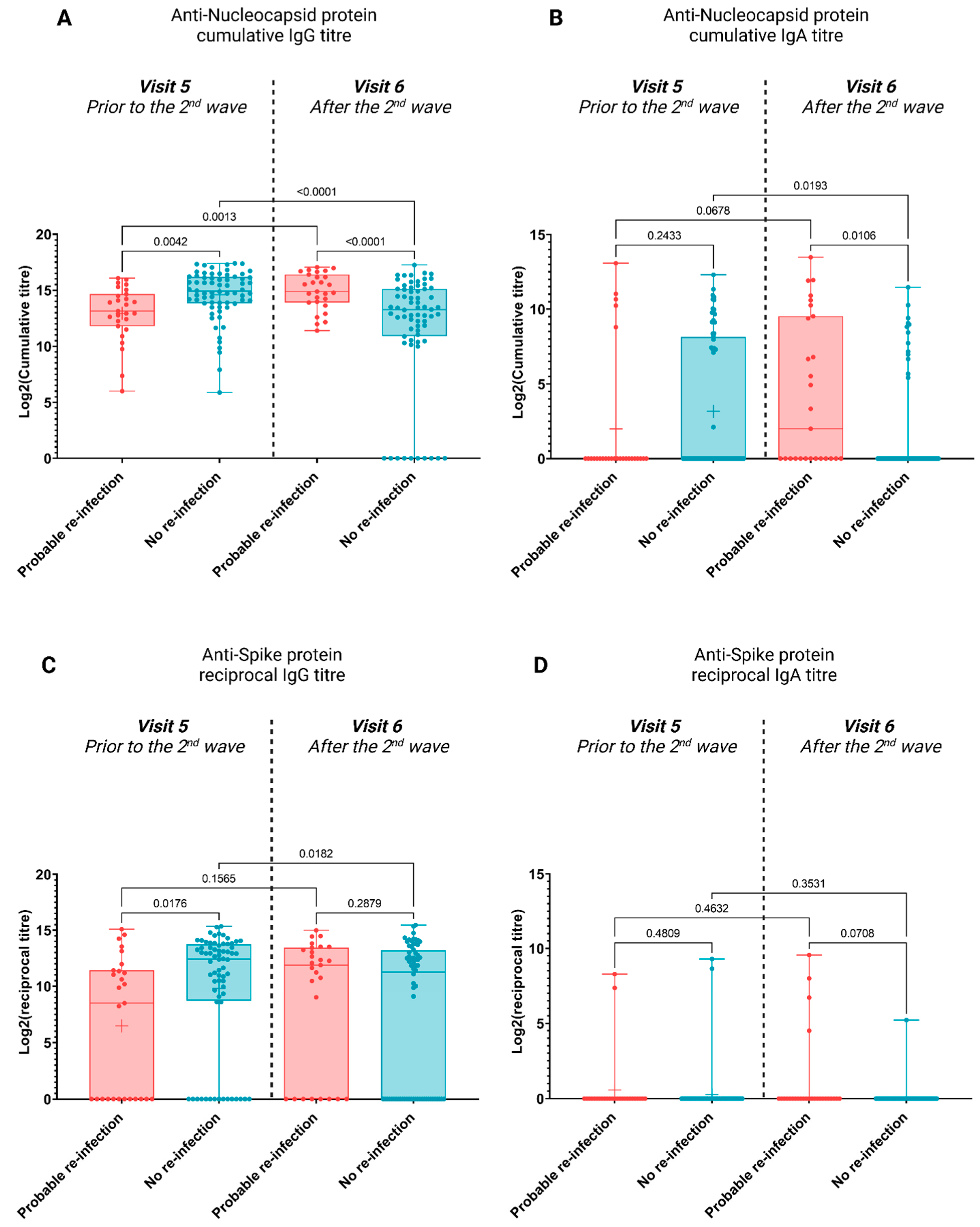
3.2. Antibody Profiles Post Potential Re-Infection
4. Discussion
Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Escobedo-de la Peña, J.; Rascón Pacheco, R.A.; de Jesús Ascencio-Montiel, I.; González-Figueroa, E.; Fernández-Gárate, J.E.; Medina-Gómez, O.S.; Borja-Bustamante, P.; Santillán-Oropeza, J.A.; Borja-Aburto, V.H. Hypertension, diabetes and obesity, major risk factors for death in patients with COVID-19 in Mexico. Arch. Med. Res. 2020, 52, 443–449. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.; Abdesselem, H.B.; Mullins, M.; Tan, T.-M.; Nel, A.J.M.; Al-Nesf, M.A.Y.; Bensmail, I.; Majbour, N.K.; Vaikath, N.N.; Naik, A.; et al. Age, Disease Severity and Ethnicity Influence Humoral Responses in a Multi-Ethnic COVID-19 Cohort. Viruses 2021, 13, 786. [Google Scholar] [CrossRef]
- Ye, C.; Zhang, S.; Zhang, X.; Cai, H.; Gu, J.; Lian, J.; Lu, Y.; Jia, H.; Hu, J.; Jin, C.; et al. Impact of comorbidities on patients with COVID-19: A large retrospective study in Zhejiang, China. J. Med. Virol. 2020, 92, 2821–2829. [Google Scholar] [CrossRef] [PubMed]
- Tillett, R.L.; Sevinsky, J.R.; Hartley, P.D.; Kerwin, H.; Crawford, N.; Gorzalski, A.; Laverdure, C.; Verma, S.C.; Rossetto, C.C.; Jackson, D.; et al. Genomic evidence for reinfection with SARS-CoV-2: A case study. Lancet Infect. Dis. 2021, 21, 52–58. [Google Scholar] [CrossRef]
- To, K.K.-W.; Hung, I.F.-N.; Ip, J.D.; Chu, A.W.-H.; Chan, W.-M.; Tam, A.R.; Fong, C.H.-Y.; Yuan, S.; Tsoi, H.-W.; Ng, A.C.-K.; et al. Coronavirus Disease 2019 (COVID-19) Re-infection by a Phylogenetically Distinct Severe Acute Respiratory Syndrome Coronavirus 2 Strain Confirmed by Whole Genome Sequencing. Clin. Infect. Dis. 2020, 73, 2946–2951. [Google Scholar] [CrossRef]
- Van Elslande, J.; Vermeersch, P.; Vandervoort, K.; Wawina-Bokalanga, T.; Vanmechelen, B.; Wollants, E.; Laenen, L.; André, E.; Van Ranst, M.; Lagrou, K.; et al. Symptomatic Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Reinfection by a Phylogenetically Distinct Strain. Clin. Infect. Dis. 2021, 73, 354–356. [Google Scholar] [CrossRef]
- West, J.; Everden, S.; Nikitas, N. A case of COVID-19 reinfection in the UK. Clin. Med. 2021, 21, e52–e53. [Google Scholar] [CrossRef]
- Brehm, T.T.; Pfefferle, S.; Von Possel, R.; Kobbe, R.; Nörz, D.; Schmiedel, S.; Grundhoff, A.; Olearo, F.; Emmerich, P.; Robitaille, A.; et al. SARS-CoV-2 Reinfection in a Healthcare Worker Despite the Presence of Detectable Neutralizing Antibodies. Viruses 2021, 13, 661. [Google Scholar] [CrossRef]
- Váncsa, S.; Dembrovszky, F.; Farkas, N.; Szakó, L.; Teutsch, B.; Bunduc, S.; Nagy, R.; Párniczky, A.; Erőss, B.; Péterfi, Z.; et al. Repeated SARS-CoV-2 Positivity: Analysis of 123 Cases. Viruses 2021, 13, 512. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Q.; Liang, Z.; Li, T.; Liu, S.; Cui, Q.; Nie, J.; Wu, Q.; Qu, X.; Huang, W.; et al. The significant immune escape of pseudotyped SARS-CoV-2 variant Omicron. Emerg. Microbes Infect. 2022, 11, 1–5. [Google Scholar] [CrossRef]
- Ibarrondo, F.J.; Fulcher, J.A.; Goodman-Meza, D.; Elliott, J.; Hofmann, C.; Hausner, M.A.; Ferbas, K.G.; Tobin, N.H.; Aldrovandi, G.M.; Yang, O.O. Rapid Decay of Anti–SARS-CoV-2 Antibodies in Persons with Mild Covid-19. N. Engl. J. Med. 2020, 383, 1085–1087. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wu, S.; Tao, H.; Zeng, G.; Zhou, F.; Guo, F.; Wang, X. Prevalence of IgG antibodies to SARS-CoV-2 in Wuhan—Implications for the ability to produce long-lasting protective antibodies against SARS-CoV-2. medRxiv 2020. [Google Scholar] [CrossRef]
- Röltgen, K.; Wirz, O.F.; Stevens, B.A.; Powell, A.E.; Hogan, C.A.; Najeeb, J.; Hunter, M.; Sahoo, M.K.; Huang, C.; Yamamoto, F.; et al. SARS-CoV-2 Antibody Responses Correlate with Resolution of RNAemia But Are Short-Lived in Patients with Mild Illness. medRxiv 2020. [Google Scholar] [CrossRef]
- Wu, L.; Wang, N.C.; Chang, Y.H.; Tian, X.Y.; Na, D.Y.; Zhang, L.Y.; Zheng, L.; Lan, T.; Wang, L.F. Duration of antibody responses after severe acute respiratory syndrome. Emerg. Infect. Dis. 2007, 13, 1562–1564. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.; Iblan, I.; Rha, B.; Alqasrawi, S.; Haddadin, A.; Al Nsour, M.; Alsanouri, T.; Ali, S.S.; Harcourt, J.; Miao, C.; et al. Persistence of Antibodies against Middle East Respiratory Syndrome Coronavirus. Emerg. Infect. Dis. 2016, 22, 1824–1826. [Google Scholar] [CrossRef]
- Blackburn, J.M.; Shoko, A.; Beeton-Kempen, N. Miniaturized, Microarray-Based Assays for Chemical Proteomic Studies of Protein Function. In Chemical Genomics and Proteomics: Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2012; Volume 800, pp. 133–162. [Google Scholar]
- Geneva: World Health Organization. WHO COVID-19 Dashboard. Available online: https://covid19.who.int/ (accessed on 8 November 2022).
- Manisty, C.; Treibel, T.A.; Jensen, M.; Semper, A.; Joy, G.; Gupta, R.K.; Cutino-Moguel, T.; Andiapen, M.; Jones, J.; Taylor, S.; et al. Time series analysis and mechanistic modelling of heterogeneity and sero-reversion in antibody responses to mild SARS-CoV-2 infection. EBioMedicine 2021, 65, 103259. [Google Scholar] [CrossRef]
- Choudhary, H.R.; Parai, D.; Dash, G.C.; Peter, A.; Sahoo, S.K.; Pattnaik, M.; Rout, U.K.; Nanda, R.R.; Pati, S.; Bhattacharya, D. IgG antibody response against nucleocapsid and spike protein post-SARS-CoV-2 infection. Infection 2021, 49, 1045–1048. [Google Scholar] [CrossRef]
- Israel, A.; Shenhar, Y.; Green, I.; Merzon, E.; Golan-Cohen, A.; Schäffer, A.A.; Ruppin, E.; Vinker, S.; Magen, E. Large-Scale Study of Antibody Titer Decay following BNT162b2 mRNA Vaccine or SARS-CoV-2 Infection. Vaccines 2021, 10, 64. [Google Scholar] [CrossRef]
- Smith, A.R.; Devies, J.; Caruso, E.; Radhakrishnan, L.; Sheppard, M.; Stein, Z.; Calanan, R.M.; Hartnett, K.P.; Kite-Powell, A.; Rodgers, L.; et al. Emergency Department Visits for COVID-19 by Race and Ethnicity—13 States, October–December 2020. MMWR. Morb. Mortal. Wkly. Rep. 2021, 70, 566–569. [Google Scholar] [CrossRef]
- Kopel, J.; Perisetti, A.; Roghani, A.; Aziz, M.; Gajendran, M.; Goyal, H. Racial and Gender-Based Differences in COVID-19. Front. Public Health 2020, 8, 418. [Google Scholar] [CrossRef]
- Knight, M.; Changrob, S.; Li, L.; Wilson, P.C. Imprinting, immunodominance, and other impediments to generating broad influenza immunity. Immunol. Rev. 2020, 296, 191–204. [Google Scholar] [CrossRef] [PubMed]
- Kubo, M.; Miyauchi, K. Breadth of Antibody Responses during Influenza Virus Infection and Vaccination. Trends Immunol. 2020, 41, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Adachi, Y.; Tonouchi, K.; Yajima, T.; Harada, Y.; Fukuyama, H.; Deno, S.; Iwakura, Y.; Yoshimura, A.; Hasegawa, H.; et al. Influenza virus infection expands the breadth of antibody responses through IL-4 signalling in B cells. Nat. Commun. 2021, 12, 3789. [Google Scholar] [CrossRef] [PubMed]

| Characteristic | ||
|---|---|---|
| Gender | Female | 220 |
| Male | 77 | |
| Not declared | 3 | |
| Age distribution | 18–30 | 89 |
| 31–40 | 80 | |
| 41–60 | 105 | |
| 61–73 | 5 | |
| Not declared | 21 | |
| Ethnicity | African | 107 |
| Caucasian | 69 | |
| Coloured | 99 | |
| Asian | 16 | |
| Other | 3 | |
| Not declared | 6 | |
| Sampling Time | Baseline | Visit 5 | Visit 6 |
|---|---|---|---|
| Seroprevalence (%) | 30.67 | 44.92 | 61.97 |
| PCR+ (%) | 14.00 | nd | nd |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mullins, M.O.; Smith, M.; Maboreke, H.; Nel, A.J.M.; Ntusi, N.A.B.; Burgers, W.A.; Blackburn, J.M. Epitope Coverage of Anti-SARS-CoV-2 Nucleocapsid IgA and IgG Antibodies Correlates with Protection against Re-Infection by New Variants in Subsequent Waves of the COVID-19 Pandemic. Viruses 2023, 15, 584. https://doi.org/10.3390/v15020584
Mullins MO, Smith M, Maboreke H, Nel AJM, Ntusi NAB, Burgers WA, Blackburn JM. Epitope Coverage of Anti-SARS-CoV-2 Nucleocapsid IgA and IgG Antibodies Correlates with Protection against Re-Infection by New Variants in Subsequent Waves of the COVID-19 Pandemic. Viruses. 2023; 15(2):584. https://doi.org/10.3390/v15020584
Chicago/Turabian StyleMullins, Michelle O., Muneerah Smith, Hazel Maboreke, Andrew J. M. Nel, Ntobeko A. B. Ntusi, Wendy A. Burgers, and Jonathan M. Blackburn. 2023. "Epitope Coverage of Anti-SARS-CoV-2 Nucleocapsid IgA and IgG Antibodies Correlates with Protection against Re-Infection by New Variants in Subsequent Waves of the COVID-19 Pandemic" Viruses 15, no. 2: 584. https://doi.org/10.3390/v15020584





