Viral Interference between the Insect-Specific Virus Brejeira and the Saint Louis Encephalitis Virus In Vitro
Abstract
:1. Introduction
2. Materials and Methods
2.1. Viral Samples
2.2. Cells
2.3. Preparation and Titration of the Viral Stock
2.4. Tissue Culture Infectious Dose 50% (TCID50)
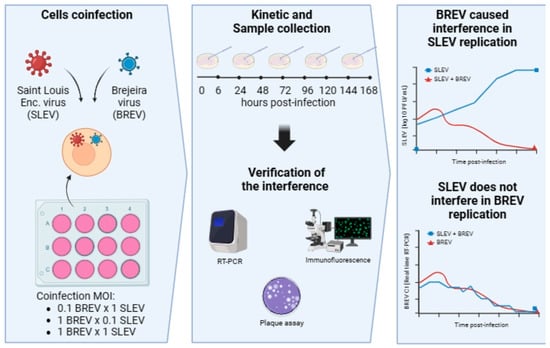
2.5. Co-Infection Experiments in Cell Culture between SLEV and BREV
2.6. Indirect Immunofluorescence (IFA) and Viral Titration
2.7. Real-Time Reverse Transcription–Polymerase Chain Reaction (Real-Time RT-PCR) for SLEV and BREV
2.8. Statistical Analysis
3. Results
3.1. Viral Titers
3.2. SLEV Co-Infection and BREV in C6/36 Cells
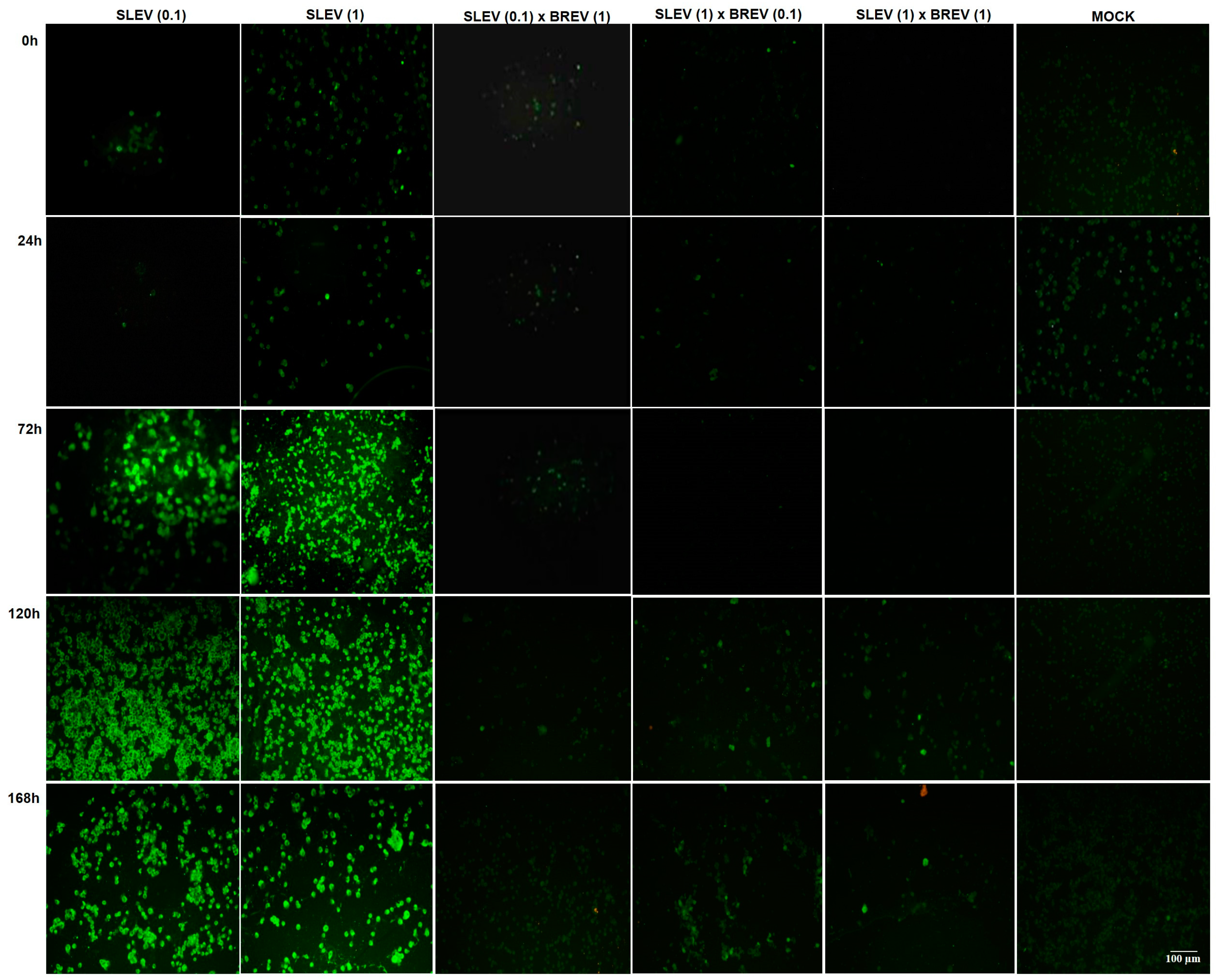
3.2.1. Observing the Cytopathic Effect (CPE)
3.2.2. Detection of SLEV Viral Antigens by IFA
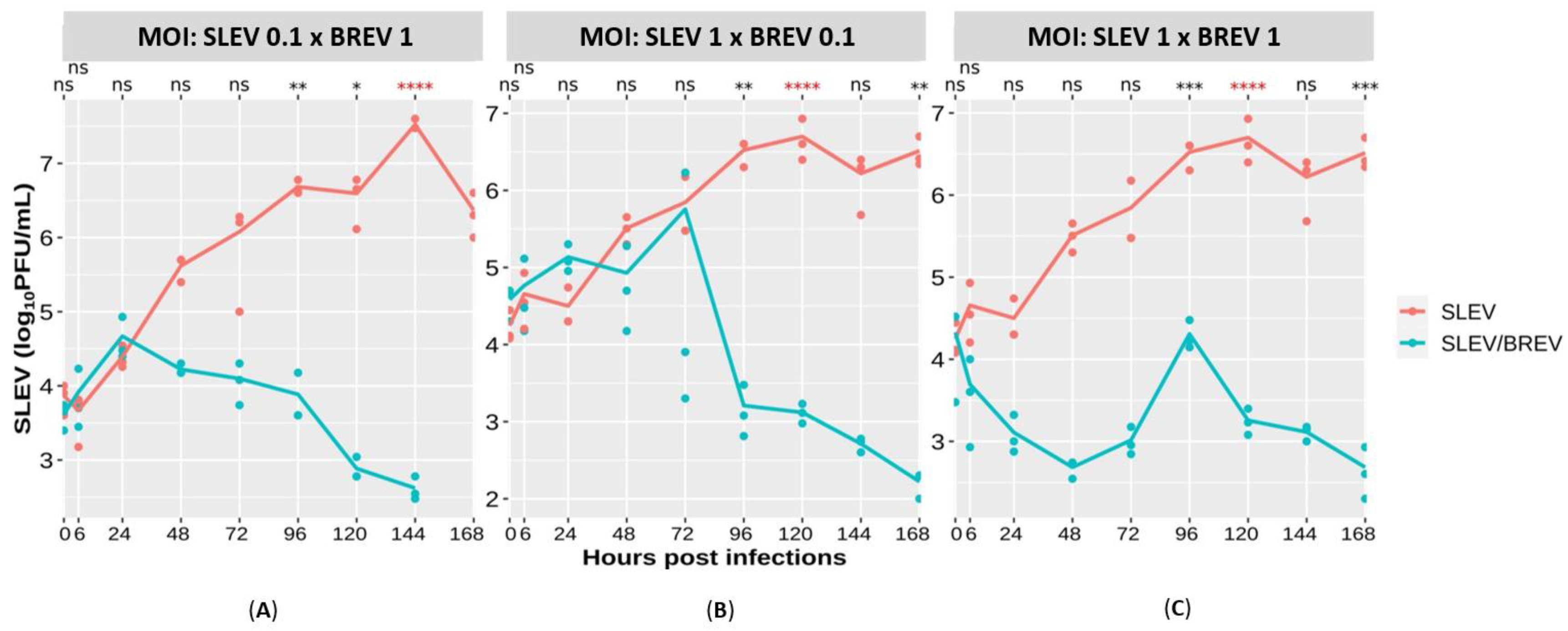
3.2.3. SLEV Titer Curve
3.2.4. Viral Genome Detection for SLEV and BREV
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vasconcelos, P.C.; Azevedo, R.S.S.; Rodrigues, S.G.; Martins, L.C.; Chiang, J.O.; Travassos Da Rosa, A.P.A. Arboviral Diseases, 1st ed.; Samauma: Belém, Brazil, 2013; Volume 1, pp. 23–574. [Google Scholar]
- Monath, T.P. Epidemiology. In St. Louis Encephalitis; American Public Health Association: Washington, DC, USA, 1980; pp. 239–312. [Google Scholar]
- Fauquet, C.M.; Mayo, J.; Maniloff, U.; Desselberger, L.; Ball, L.A. Vírus Taxonomy: Classification and Nomenclature of Viroses; Academic Press: Amsterdam, The Netherland, 2005; p. 1259. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). SLEV. 2023. Available online: https://www.cdc.gov/sle/ (accessed on 24 November 2023).
- International Committee on Taxonomy of Viruses (ICTV). 2023. Available online: https://ictv.global/report/chapter/flaviviridae/flaviviridae (accessed on 24 November 2023).
- Leake, J.P. Epidemiology of Encephalitis: With Special Reference to the 1933 Epidemic. Am. J. Public Health Nations Health 1933, 23, 1140–1143. [Google Scholar] [CrossRef] [PubMed]
- Muckenfuss, R.S. Clinical Observations and Laboratory Investigations on the 1933 Epidemic of Encephalitis in St. Louis. Bull. N. Y. Acad. Med. 1934, 10, 444–453. [Google Scholar]
- Rodrigues, S.G.; Oliva, O.P.; Araujo, F.A.A.; Martins, L.C.; Chiang, J.O.; Henriques, D.F.; Silva, E.V.P.; Rodrigues, D.S.G.; Prazeres, A.S.C.; Tavares Neto, J.; et al. Epidemiology of the Saint Louis encephalitis virus. Rev. Pan-Amaz. Health 2010, 1, 81–86. [Google Scholar] [CrossRef]
- Heinen, L.B.S.; Zuchi, N.; Serra, O.P.; Cardoso, B.F.; Gondim, B.H.F.; Santos, M.A.M.; Souto, F.J.D.; Paula, D.A.J.; Dutra, V.; Dezengrini-Slhessarenko, R. Saint Louis encephalitis virus in Mato Grosso, central-western Brazil. Inst. Trop. Med. São Paulo 2015, 57, 215–220. [Google Scholar] [CrossRef]
- Vasconcelos, P.C.; Travassos da Rosa, J.F.S.; Travassos da Rosa, A.P.A.; Degallier, N. Epidemiology of Arbovirus Encephalitis in the Brazilian Amazon. Inst. Trop. Med. São Paulo 1991, 3, 465–476. [Google Scholar] [CrossRef]
- Travassos da Rosa, A.P.A.; Travassos da Rosa, J.F.S.; Pinheiro, F.P.; Vasconcelos, P.C. Arboviroses. In Focus on Infectious and Parasitic Diseases in the Amazon; CEJUP: Belém, Brazil, 1997; pp. 207–225. [Google Scholar]
- Diaz, A.; Coffey, L.L.; Cadena, N.B.; Day, J.F. Reemergence of St. Louis Encephalitis Vírus in the Americas. Emerg. Infect. Dis. 2018, 24, 2150–2157. [Google Scholar] [CrossRef]
- Reisen, W.K. Epidemiology of St. Louis encephalitis vírus. In Advances in Vírus Research; Maramorosch, K., Murphy, F.A., Shatkin, A.J., Eds.; Academic Press: San Diego, CA, USA, 2003; Volume 61, pp. 139–183. [Google Scholar]
- Brinker, K.R.; Monath, T.P. The acute disease. In Saint Louis Encephalitis; American Public Health Association: Washington, DC, USA, 1980; pp. 503–534. [Google Scholar]
- Bolling, B.G.; Vasilakis, N.; Guzman, H.; Widen, S.G.; Wood, T.G.; Popov, V.L.; Thangamani, S.; Tesh, R.B. Insect-Specific Víruses Detected in Laboratory Mosquito Colonies and Their Potential Implications for Experiments Evaluating Arbovírus Vector Competence. Am. J. Trop. Med. 2015, 92, 422–428. [Google Scholar] [CrossRef] [PubMed]
- Vasilakis, N.; Forrester, N.L.; Palacios, G.; Nasar, F.; Savjid, N.; Rossi, S.L.; Guzman, H.; Wood, T.G.; Popov, V.; Gorchakov, R.; et al. Negevírus: A proposed new taxon of insect-specific víruses with wide geographic distribution. J. Virol. 2013, 87, 2475–2488. [Google Scholar] [CrossRef]
- Vasilakis, N.; Tesh, R.B. Insect-specific víruses and their potential impact on arbovírus transmission. Curr. Opin. Virol. 2015, 15, 69–74. [Google Scholar] [CrossRef]
- Kallies, R.; Kopp, A.; Zirkel, F.; Estrada, A.; Gillespie, T.R.; Drosten, C.; Junglen, S. Genetic characterization of Goutanap vírus, a novel vírus related to Negevíruses, cilevíruses and higrevíruses. Víruses 2014, 6, 4346–4357. [Google Scholar] [CrossRef]
- Nunes, M.R.T.; Contreras-Gutierrez, M.A.; Guzman, H.; Martins, L.C.; Barbirato, M.F.; Savit, C.; Balta, V.; Uribe, S.; Vivero, R.; Suaza, J.D.; et al. Genetic characterization, molecular epidemiology, and phylogenetic relationships of insect-specificviruses in the taxon Negevírus. Virology 2017, 504, 152–167. [Google Scholar] [CrossRef]
- Nunes, M.R.T.; Silva, S.P.; Carvalho, V.L.; Vasconcelos, J.M.; Silva, D.E.A.; Oliveira, L.F.; Nunes Neto, J.P.; Rodrigues, S.G.; Azevedo, R.S.S.; Monteiro, H.A.O.; et al. Emergence of new insect-restrictive víruses in the Amazon region. Genome Announc. 2015, 2, e00131-15. [Google Scholar] [CrossRef] [PubMed]
- Ribeiro, A.C.S.; Silva, F.S.; Gomes, F.P.; Barros, L.J.L.; Casseb, L.M.N.; Martins, L.C.; Carvalho, V.L. Insect-specific viruses of the negevirus taxon: Analysis of the characteristics of Brejeira virus. Electron. Mag. Acervo Saúde 2023, 23, e13686. [Google Scholar] [CrossRef]
- Ribeiro, A.C.S.; Martins, L.C.; Silva, S.P.; Medeiros, D.B.A.; Miranda, K.K.P.; Nunes Neto, J.P.; Monteiro, H.A.D.O.; Nascimento, B.L.S.; Rosa Junior, J.W.; Cruz, A.C.R.; et al. Negeviruses isolated from mosquitoes in the Brazilian Amazon. Virol. J. 2022, 19, 17. [Google Scholar] [CrossRef]
- Baidaliuk, A.; Miot, E.F.; Lequime, S.; Moltini-Conclois, I.; Delaigue, F.; Dabo, S.; Dickson, L.B.; Aubry, F.; Merkling, S.H.; Cao-Lormeau, V.M.; et al. Cell-Fusing Agent Virus Reduces Arbovirus Dissemination in Aedes aegypti Mosquitoes In Vivo. J. Virol. 2019, 93, e00705-19. [Google Scholar] [CrossRef]
- Goenaga, S.; Kenney, J.L.; Duggal, N.K.; Delorey, M.; Ebel, G.D.; Zhang, B.; Levis, S.C.; Enria, D.A.; Brault, A.C. Potential for Co-Infection of a Mosquito-Specific Flavivirus, Nhumirim Virus, to Block West Nile virus Transmission in Mosquitoes. Viruses 2015, 7, 5801–5812. [Google Scholar] [CrossRef]
- Gorchakov, R.V.; Tesh, R.B.; Weaver, S.C.; Nasar, F. Generation of an infectious Negev virus cDNA clone. J. Gen. Virol. 2014, 9, 2071. [Google Scholar] [CrossRef] [PubMed]
- Erasmus, J.H.; Weaver, S.C. Biotechnological applications of an insect-specific alphavirus. DNA Cell Biol. 2017, 12, 1045–1049. [Google Scholar] [CrossRef] [PubMed]
- Erasmus, J.H.; Auguste, A.J.; Kaelber, J.T.; Luo, H.; Rossi, S.L.; Fenton, K.; Leal, G.; Kim, D.Y.; Chiu, W.; Wang, T.; et al. A chikungunya fever vaccine utilizing an insect-specific virus platform. Nat. Med. 2017, 2, 192–199. [Google Scholar] [CrossRef]
- Patterson, E.; Kautz, T.F.; Contreras-Gutierrez, M.A.; Guzman, H.; Tesh, R.B.; Hughes, G.L.; Forrester, N.L. Negevíruses reduce replication of alfavíruses during coinfection. J. Virol. 2020, 95, e0043321. [Google Scholar] [CrossRef]
- Igarashi, A. Isolation of a Singh’s Aedes albopictus cell clone sensitive to Dengue and Chikungunya viruses. J. Gen. Virol. 1978, 40, 531–544. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, M.L.; Rocco, I.M.; Felippe, J.M.M.S.; Cruz, A.S. Growth and maintenance of Aedes Albopictus cell line, clone C6/36, in different media. Rev. Do Inst. Adolf Lutz. 1993, 53, 63–70. [Google Scholar]
- Ammerman, N.C.; Beier-Sexton, M.; Azad, A.F. Growth and Maintenance of Vero Cell Lines. Curr. Protoc. Microbiol. 2008, 11, A-4E. [Google Scholar] [CrossRef] [PubMed]
- Malewicz, B.; Jenkin, H.M. Development of dengue vírus plaques under serum-free overlay medium. J. Clin. Microbiol. 1979, 9, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Reed, L.J.; Muench, H. A Simple Method Of Estimating Fifty Per Cent Endpoints American. J. Epidemiol. 1938, 27, 493–497. [Google Scholar] [CrossRef]
- ATCC. ATCC Virology Culture Guide. Available online: https://www.atcc.org/resources/culture-guides/virology-culture-guide#:~:text=For%20any%20titer%20expressed%20as,%C3%97%20105%20PFU%2FmL (accessed on 20 January 2024).
- Gubler, D.J.; Kuno, G.; Sather, G.E.; Oliver, V.L.A. Mosquito Cell Cultures and Specific Monoclonal Antibodies in Surveillance for Dengue Víruses. Am. J. Trop. Med. Hyg. 1984, 33, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Brackney, D.E.; Scott, J.C.; Sagawa, F.; Woodward, J.E.; Miller, N.A.; Schilkey, F.D.; Mudge, J.; Wilusz, J.; Olson, K.E.; Blair, C.D.; et al. C6/36 Aedes albopictus Cells Have a Dysfunctional Antiviral RNA Interference Response. PLoS Negl. Trop. Dis. 2010, 4, e856. [Google Scholar] [CrossRef]
- Blair, C.D. Mosquito RNAi is the major innate immune pathway controlling arbovírus infection and transmission. Future Microbiol. 2011, 6, 265–277. [Google Scholar] [CrossRef]
- Romo, H.; Kenney, J.L.; Blitvich, B.J.; Aaron, C.B. Restriction of Zika vírus infection and transmission in Aedes aegypti mediated by an insect-specific flavivírus. Emerg. Microbes Infect. 2018, 7, 181. [Google Scholar] [CrossRef]
- Schultz, M.J.; Frydman, H.M.; Connor, J.H. Dual Insect specific virus infection limits Arbovirus replication in Aedes mosquito cells. Virology 2018, 518, 406–413. [Google Scholar] [CrossRef]
- Hall, R.A.; Helle, O.B.; Breeanna, J.M.; O’brien, C.A.; Colmant, A.M.G.; Piyasena, T.B.H.; Harrison, J.J.; Newton, N.D.; Barnard, R.T.; Natalie, A.P.; et al. Commensal Víruses of Mosquitoes: Host Restriction, Transmission, and Interaction with Arboviral Pathogens. Evol. Bioinform. 2016, 12, 35–44. [Google Scholar] [CrossRef] [PubMed]
- Hobson-Peters, J.; Yam, A.W.Y.; Lu, J.W.F.; Setoh, Y.X.; May, F.J.; Kurucz, N.; Walsh, S.; Prow, N.A.; Davis, S.S.; Weir, R.; et al. A new insect-specific flavivirus from northern Australia suppresses replication of West Nile virus and Murray valley encephalitis virus in co-infected mosquito cells. PLoS ONE 2013, 8, e56534. [Google Scholar] [CrossRef]


| Virus | Primers/Probes |
|---|---|
| SLEV | SLE2420 2420–2439 CTGGCTGTCGGAGGGATTCT 68 (Primer) |
| SLE2487c 2487–2468 TAGGTCAATTGCACATCCCG (Primer) | |
| SLE2444-probe 2444–2466 TCTGGCGACCAGCGTGCAAGCCG (Probe) | |
| SLE834 834–852 GAAAACTGGGTTCTGCGCA 72 (Primer) | |
| SLE905c 905–889 GTTGCTGCCTAGCATCCATCC (Primer) | |
| SLE857-probe 857–880 TGGATATGCCCTAGTTGCGCTGGC (Probe) | |
| BREV | BREV–F–ATGACCGATGATGAGAACCG (Primer) |
| BREV–R–GGTGAGACAGCAATAGTAGCC (Primer) | |
| BREV–Probe–TCGTGCTCGATGACACCCGC (Probe) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ribeiro, A.C.; Martins, L.; Silva, H.; Freitas, M.N.; Santos, M.; Gonçalves, E.; Sousa, A.; Prazeres, I.; Santos, A.; Cruz, A.C.; et al. Viral Interference between the Insect-Specific Virus Brejeira and the Saint Louis Encephalitis Virus In Vitro. Viruses 2024, 16, 210. https://doi.org/10.3390/v16020210
Ribeiro AC, Martins L, Silva H, Freitas MN, Santos M, Gonçalves E, Sousa A, Prazeres I, Santos A, Cruz AC, et al. Viral Interference between the Insect-Specific Virus Brejeira and the Saint Louis Encephalitis Virus In Vitro. Viruses. 2024; 16(2):210. https://doi.org/10.3390/v16020210
Chicago/Turabian StyleRibeiro, Ana Cláudia, Lívia Martins, Heloisa Silva, Maria Nazaré Freitas, Maissa Santos, Ercília Gonçalves, Alana Sousa, Ivy Prazeres, Alessandra Santos, Ana Cecilia Cruz, and et al. 2024. "Viral Interference between the Insect-Specific Virus Brejeira and the Saint Louis Encephalitis Virus In Vitro" Viruses 16, no. 2: 210. https://doi.org/10.3390/v16020210