Characterizing Infections in Two Epidemic Waves of SARS-CoV-2 Omicron Variants: A Cohort Study in Guangzhou, China
Abstract
:1. Introduction
2. Materials and Methods
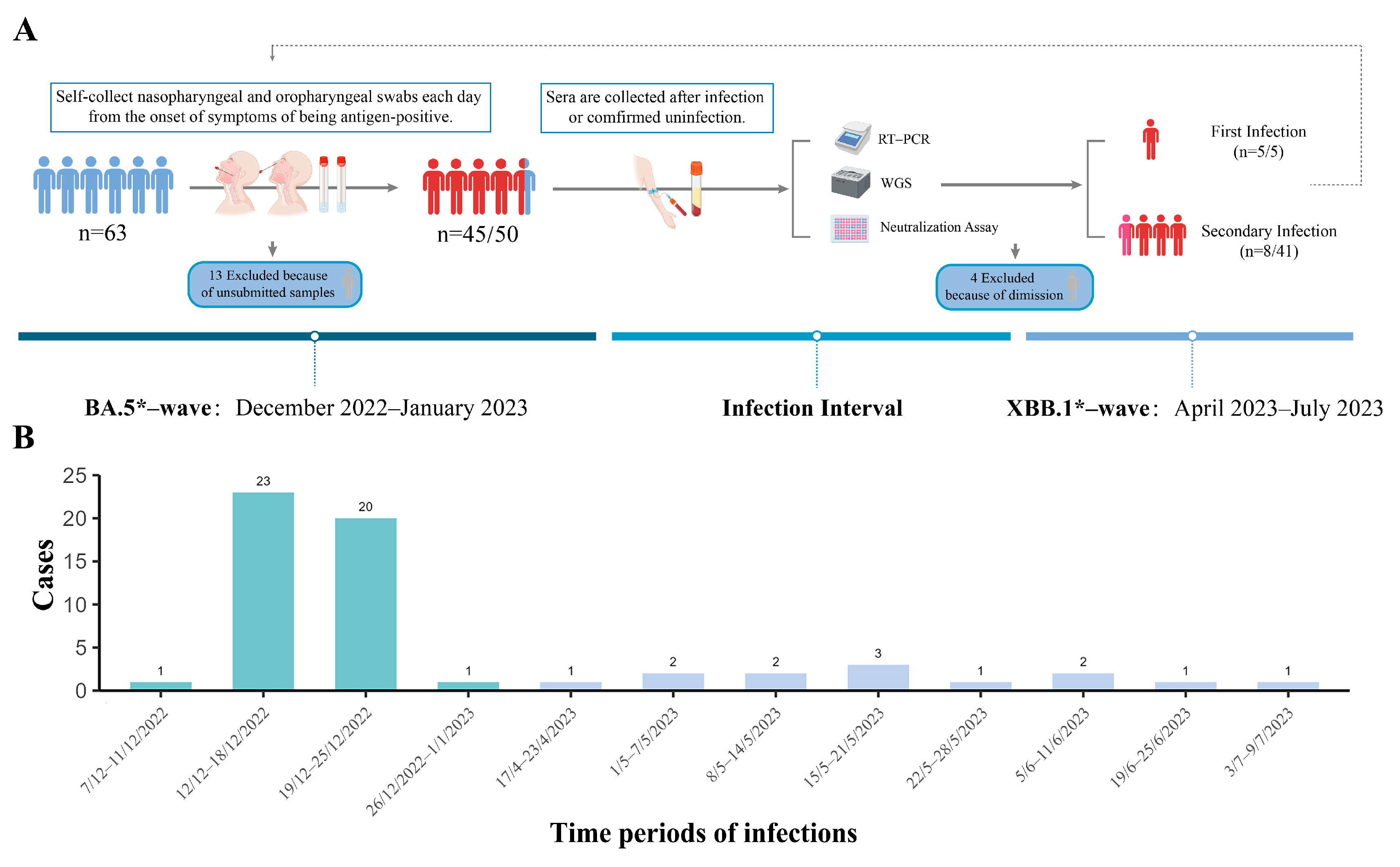
2.1. Study Design and Participants
2.2. RT-PCR
2.3. Whole Genome Sequencing, WGS
2.4. Phylogenetic Analysis
2.5. Microneutralization Detection
2.6. Statistical Analysis
3. Results
3.1. Epidemiology Characters
3.2. Viral Trajectories of Different Variant Infections in Various Sample Types
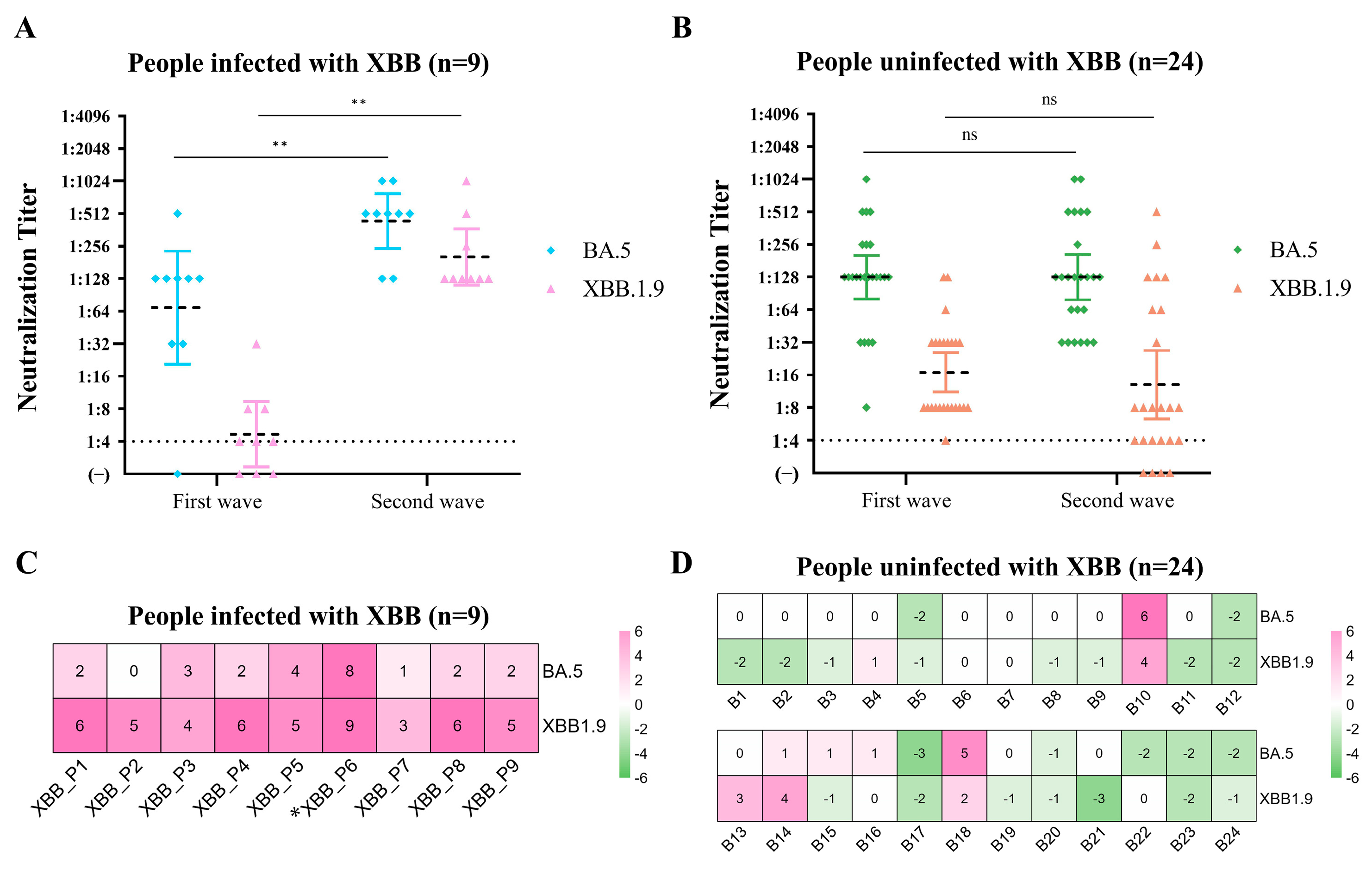
3.3. Changes in Convalescent Sera-Neutralizing Antibodies in the Cohort Population after the Two Epidemics
4. Discussion
Limitation of the Study
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- WHO Coronavirus (COVID-19) Dashboard 2023. Available online: https://covid19.who.int (accessed on 5 December 2023).
- Mistry, P.; Barmania, F.; Mellet, J.; Peta, K.; Strydom, A.; Viljoen, I.M.; James, W.; Gordon, S.; Pepper, M.S. SARS-CoV-2 Variants, Vaccines, and Host Immunity. Front. Immunol. 2021, 12, 809244. [Google Scholar] [CrossRef]
- Vitiello, A.; Ferrara, F.; Auti, A.M.; Domenico, M.D.; Boccellino, M. Advances in the Omicron variant development. J. Intern. Med. 2022, 292, 81–90. [Google Scholar] [CrossRef]
- Hu, Z.; Jin, Z.; Zhou, M.; Zhang, C.; Bao, Y.; Gao, X.; Wang, G. CoronaVac and BBIBP-CorV vaccines against SARS-CoV-2 during predominant circulation of Omicron BA.5.2 and BF.7 in China, a retrospective cohort study. J. Med. Virol. 2023, 95, e29143. [Google Scholar] [CrossRef]
- Cai, J.; Deng, X.; Yang, J.; Sun, K.; Liu, H.; Chen, Z.; Peng, C.; Chen, X.; Wu, Q.; Zou, J.; et al. Modeling transmission of SARS-CoV-2 Omicron in China. Nat. Med. 2022, 28, 1468–1475. [Google Scholar] [CrossRef]
- The National Health Commission of People’s Republic of China. Control Mechanism for COVID-19 under the State Council of P.R. China on December 07. Prevention and control measures to further optimize COVID-19 response. Health Care Sci. 2023, 2, 7–9. [Google Scholar] [CrossRef]
- Chinese Center for Disease Control and Prevention 2023. Available online: https://www.chinacdc.cn/jkzt/crb/zl/szkb_11803/jszl_12208/202301/t20230114_263378.html (accessed on 5 December 2023).
- Abdullah, F.; Myers, J.; Basu, D.; Tintinger, G.; Ueckermann, V.; Mathebula, M.; Ramlall, R.; Spoor, S.; de Villiers, T.; Van der Walt, Z.; et al. Decreased severity of disease during the first global omicron variant covid-19 outbreak in a large hospital in tshwane, south africa. Int. J. Infect. Dis. 2022, 116, 38–42. [Google Scholar] [CrossRef]
- Li, B.; Deng, A.; Li, K.; Hu, Y.; Li, Z.; Shi, Y.; Xiong, Q.; Liu, Z.; Guo, Q.; Zou, L.; et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat. Commun. 2022, 13, 460. [Google Scholar] [CrossRef]
- Barcoding (SQK-RBK004)-MinION-Overview of the Protocol. Available online: https://community.nanoporetech.com/docs/prepare/library_prep_protocols/rapid-barcoding-sequencing-sqk-rbk004/v/rbk_9054_v2_revaf_14aug2019/overview-of-the-rapid-barc?devices=minion (accessed on 5 December 2023).
- Hadfield, J.; Megill, C.; Bell, S.M.; Huddleston, J.; Potter, B.; Callender, C.; Sagulenko, P.; Bedford, T.; Neher, R.A. Nextstrain: Real-time tracking of pathogen evolution. Bioinformatics 2018, 34, 4121–4123. [Google Scholar] [CrossRef]
- Sun, J.; Tang, X.; Bai, R.; Liang, C.; Zeng, L.; Lin, H.; Yuan, R.; Zhou, P.; Huang, X.; Xiong, Q.; et al. The kinetics of viral load and antibodies to SARS-CoV-2. Clin. Microbiol. Infect. 2020, 26, 1690.e1–1690.e4. [Google Scholar] [CrossRef]
- The Novel Coronavirus Pneumonia Emergency Response Epidemiology Team. The Epidemiological Characteristics of an Outbreak of 2019 Novel Coronavirus Diseases (COVID-19)—China, 2020. China CDC Wkly. 2020, 2, 113–122. [Google Scholar] [CrossRef]
- Niu, B.; Ji, S.; Zhao, S.; Lei, H. Timing and Magnitude of the Second Wave of the COVID-19 Omicron Variant—189 Countries and Territories, November 2021 to February 2023. China CDC Wkly. 2023, 5, 397–401. [Google Scholar] [CrossRef]
- Ouyang, F.; Zhang, H.; Jiang, Y.; Wang, H.; Peng, T.; Xi, C.; Yu, J.; Li, Z.; Chen, J.; Wu, L.; et al. Humoral immune response characteristics of the elderly, children and pregnant women after XBB infection. J. Infect. 2023, 87, e96–e99. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Peng, Z.; Wei, F.; Jin, Z.; Wang, J.; Xu, X.; Zhang, X.; Xu, J.; Ren, Z.; Lu, B.; et al. Study on the COVID-19 epidemic in mainland China between November 2022 and January 2023, with prediction of its tendency. J. Biosaf. Biosecur. 2023, 5, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Kissler, S.M.; Fauver, J.R.; Mack, C.; Tai, C.G.; Breban, M.I.; Watkins, A.E.; Samant, R.M.; Anderson, D.J.; Metti, J.; Khullar, G.; et al. Viral Dynamics of SARS-CoV-2 Variants in Vaccinated and Unvaccinated Persons. N. Engl. J. Med. 2021, 385, 2489–2491. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Huang, J.; Yu, Y.; Wan, Z.; Chiu, M.C.; Liu, X.; Zhang, S.; Cai, J.-P.; Chu, H.; Li, G.; et al. Human airway and nasal organoids reveal escalating replicative fitness of SARS-CoV-2 emerging variants. Proc. Natl. Acad. Sci. USA 2023, 120, e2300376120. [Google Scholar] [CrossRef] [PubMed]
- Boroojerdi, M.H.; Al Jabry, T.; Mirarefin, S.M.J.; Albalushi, H. Insights into organoid-based modeling of COVID-19 pathology. Virol. J. 2023, 20, 37. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.; Hu, B.; Zhao, X.; Chi, H.; Pan, J.; Zheng, Y.; Bi, X.; Chen, M.; Xie, J.; Xu, J.; et al. Longitudinal observation of viral load in patients infected with Omicron variant and its relationship with clinical symptoms. Front. Microbiol. 2022, 13, 1037733. [Google Scholar] [CrossRef]
- Li, K.; Wang, D.; Qu, H.; Rui, J.; Abudunaibi, B.; Guo, Z.; Wu, S.; Abudurusuli, G.; Yang, Z.; Fang, K.; et al. Viral dynamics of omicron BA.2.76 variant of SARS-CoV-2 in a cohort of COVID-19 patients. J. Infect. 2023, 86, 154–225. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Ishikane, M.; Ujiie, M.; Iwamoto, N.; Okumura, N.; Sato, T.; Nagashima, M.; Moriya, A.; Suzuki, M.; Hojo, M.; et al. Duration of Infectious Virus Shedding by SARS-CoV-2 Omicron Variant-Infected Vaccinees. Emerg. Infect. Dis. 2022, 28, 998–1001. [Google Scholar] [CrossRef]
- Puhach, O.; Meyer, B.; Eckerle, I. SARS-CoV-2 viral load and shedding kinetics. Nat. Rev. Microbiol. 2023, 21, 147–161. [Google Scholar] [CrossRef]
- Nasal Vaccine Could Be New Weapon against COVID-19 2023. Available online: https://www.nature.com/articles/d42473-023-00095-8 (accessed on 5 December 2023).
- Chen, J.; Wang, P.; Yuan, L.; Zhang, L.; Zhang, L.; Zhao, H.; Chen, C.; Wang, X.; Han, J.; Chen, Y.; et al. A live attenuated virus-based intranasal COVID-19 vaccine provides rapid, prolonged, and broad protection against SARS-CoV-2. Sci. Bull. 2022, 67, 1372–1387. [Google Scholar] [CrossRef] [PubMed]
- Waltz, E. How nasal-spray vaccines could change the pandemic. Nature 2022, 609, 240–242. [Google Scholar] [CrossRef]
- COVID-19 Vaccine Tracker and Landscape 2023. Available online: https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines (accessed on 5 December 2023).
- Yue, C.; Song, W.; Wang, L.; Jian, F.; Chen, X.; Gao, F.; Shen, Z.; Wang, Y.; Wang, X.; Cao, Y.; et al. ACE2 binding and antibody evasion in enhanced transmissibility of XBB.1.5. Lancet Infect. Dis. 2023, 23, 278–280. [Google Scholar] [CrossRef] [PubMed]
- Regan, J.J.; Moulia, D.L.; Link-Gelles, R.; Godfrey, M.; Mak, J.; Najdowski, M.; Rosenblum, H.G.; Shah, M.M.; Twentyman, E.; Meyer, S.; et al. Use of Updated COVID-19 Vaccines 2023-2024 Formula for Persons Aged ≥ 6 Months: Recommendations of the Advisory Committee on Immunization Practices - United States, September 2023. MMWR Morb. Mortal. Wkly. Rep. 2023, 72, 1140–1146. [Google Scholar] [CrossRef] [PubMed]
- Statement on the Fifteenth Meeting of the IHR (2005) Emergency Committee on the COVID-19 Pandemic. Available online: https://www.who.int/news/item/05-05-2023-statement-on-the-fifteenth-meeting-of-the-international-health-regulations-(2005)-emergency-committee-regarding-the-coronavirus-disease-(covid-19)-pandemic (accessed on 5 December 2023).
- Xu, X.; Sun, J.; Nie, S.; Li, H.; Kong, Y.; Liang, M.; Hou, J.; Huang, X.; Li, D.; Ma, T.; et al. Seroprevalence of immunoglobulin M and G antibodies against SARS-CoV-2 in China. Nat. Med. 2020, 26, 1193–1195. [Google Scholar] [CrossRef]
- Outbreak.info SARS-CoV-2 Data Explorer 2023. Available online: https://outbreak.info/ (accessed on 5 December 2023).


| Characteristics | BA.5*-Wave (n = 45) No. (%) | XBB.1*-Wave (n = 13) No. (%) | p Value |
|---|---|---|---|
| Age, Mean ± standard deviation | 32.51 ± 8.54 | 29.00 ± 5.22 | |
| Sex | |||
| M | 16 (42.22) | 5 (38.46) | |
| F | 29 (64.44) | 8 (61.54) | |
| Variant | |||
| Omicron BA.5 variants | |||
| BA.5.2 | 1 (2.22) | 0 (0.00) | |
| BA.5.2.49 | 1(2.22) | 0 (0.00) | |
| DY.3 | 43 (95.56) | 0 (0.00) | |
| Omicron XBB.1 variants | |||
| FL.2.3 | 0 (0.00) | 7 (53.85.69) | |
| FU.1 | 0 (0.00) | 1 (7.69) | |
| EG.5.1.1 | 0 (0.00) | 1 (7.69) | |
| XBB.1.16 | 0 (0.00) | 2 (15.38) | |
| FL.15 | 0 (0.00) | 1 (7.69) | |
| BN.1.2.6 | 0 (0.00) | 1 (7.69) | |
| Months between infection and the last dose of vaccination, Median (IQR) | 13.77 (11.92,14.02) | 17.53 (15.32,19.22) | |
| Symptoms when diagnosed with COVID-19 | |||
| Fever (>37.3 °C) | 44 (97.78) | 7 (53.85) | <0.001 |
| Cough | 33 (73.33) | 8 (61.54) | >0.05 |
| Asthenia | 39 (86.67) | 3 (23.08) | <0.001 |
| Anosmia or taste abnormality | 10 (22.22) | 4 (30.77) | >0.05 |
| Nasal obstruction | 26 (57.78) | 3 (23.08) | >0.05 |
| Rhinorrhea | 31 (68.89) | 5 (38.46) | >0.05 |
| Sore throat | 34 (75.56) | 9 (69.23) | >0.05 |
| Conjunctivitis | 1 (2.22) | 0 (0.00) | >0.05 * |
| Myalgia | 31 (68.89) | 4 (30.77) | <0.05 |
| Diarrhea | 4 (8.89) | 1 (7.69) | >0.05 |
| Rash | 0 (0.00) | 3 (23.08) | <0.01 * |
| Headaches | 0 (0.00) | 3 (23.08) | <0.01 * |
| Respiratory sample tested | |||
| Nasopharyngeal swab | 281 | 76 | |
| Oropharyngeal swab | 298 | 77 | |
| Sampling days (Mean ± standard deviation) | 6.69 ± 1.28 | 5.92 ± 2.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qu, L.; Xie, C.; Qiu, M.; Yi, L.; Liu, Z.; Zou, L.; Hu, P.; Jiang, H.; Lian, H.; Yang, M.; et al. Characterizing Infections in Two Epidemic Waves of SARS-CoV-2 Omicron Variants: A Cohort Study in Guangzhou, China. Viruses 2024, 16, 649. https://doi.org/10.3390/v16040649
Qu L, Xie C, Qiu M, Yi L, Liu Z, Zou L, Hu P, Jiang H, Lian H, Yang M, et al. Characterizing Infections in Two Epidemic Waves of SARS-CoV-2 Omicron Variants: A Cohort Study in Guangzhou, China. Viruses. 2024; 16(4):649. https://doi.org/10.3390/v16040649
Chicago/Turabian StyleQu, Lin, Chunyan Xie, Ming Qiu, Lina Yi, Zhe Liu, Lirong Zou, Pei Hu, Huimin Jiang, Huimin Lian, Mingda Yang, and et al. 2024. "Characterizing Infections in Two Epidemic Waves of SARS-CoV-2 Omicron Variants: A Cohort Study in Guangzhou, China" Viruses 16, no. 4: 649. https://doi.org/10.3390/v16040649





