Deletion of 82–85 N-Terminal Residues in SARS-CoV-2 Nsp1 Restricts Virus Replication
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Cultures and Virus Isolates
2.2. Plasmids
2.3. Kinetics of Viral Replication
2.4. Assessment of IFN-β Expression
2.5. Western Blotting
2.6. Reverse-Transcription Quantitative PCR (RT-qPCR)
2.7. Cycloheximide Chase Analysis and NSs Protein Stability
2.8. Computational Details
2.9. Statistics
3. Results
3.1. SARS-CoV-2 Strains Bearing Nsp1 N-Terminal Deletions Are Attenuated in Replication
3.2. INF-β Production by Nsp1 Deletion Strains According to Different Growth Kinetics
3.3. The N-Terminal Region of Nsp1 Emerges as Crucial for Its Interaction with the Host Cellular Transcription/Translation Machinery
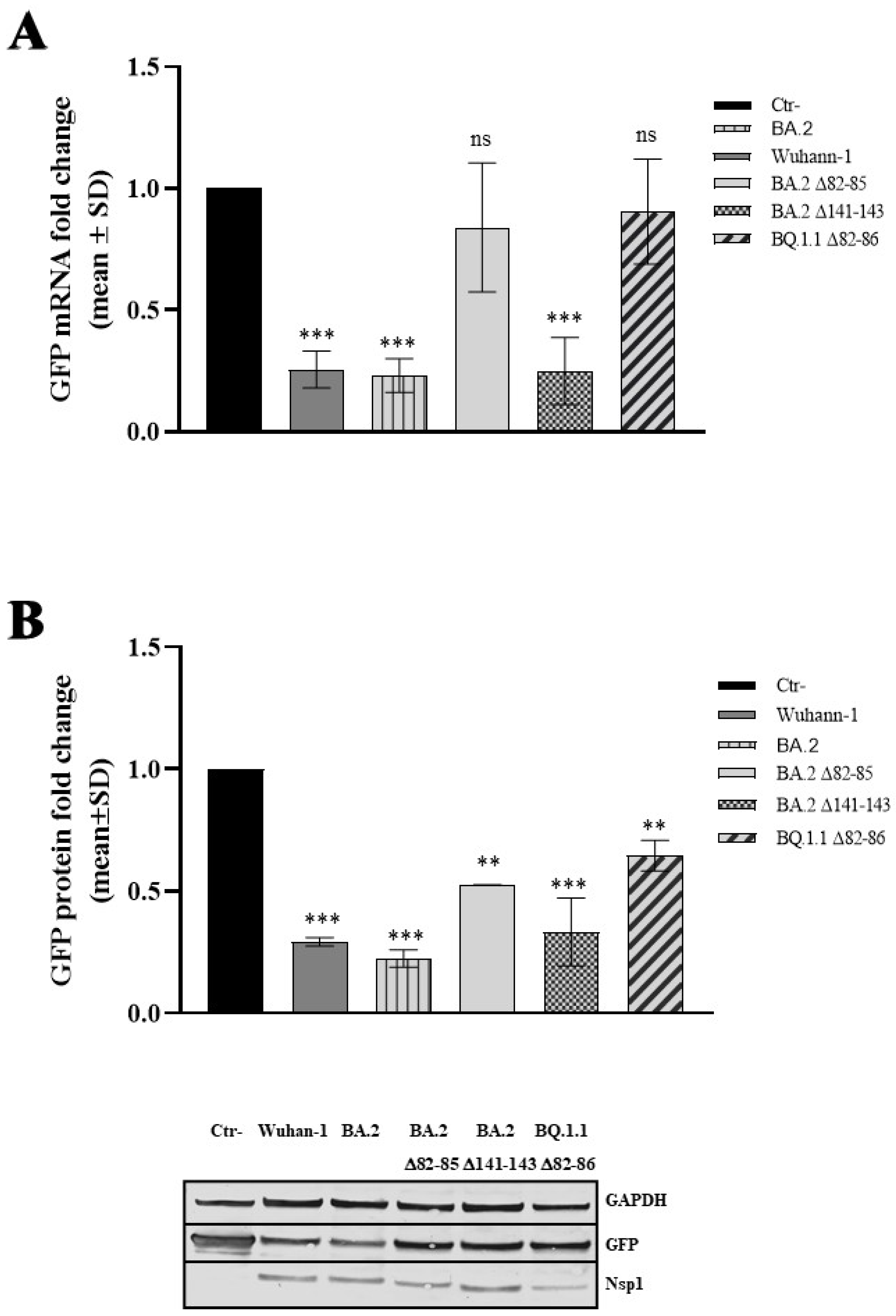
3.4. The 82–85 Nsp1 N-Terminal Domain Is Involved in Viral mRNA Expression
3.5. The 82–85 Nsp1 Domain Is Required for Cellular and Viral mRNAs Stability Control
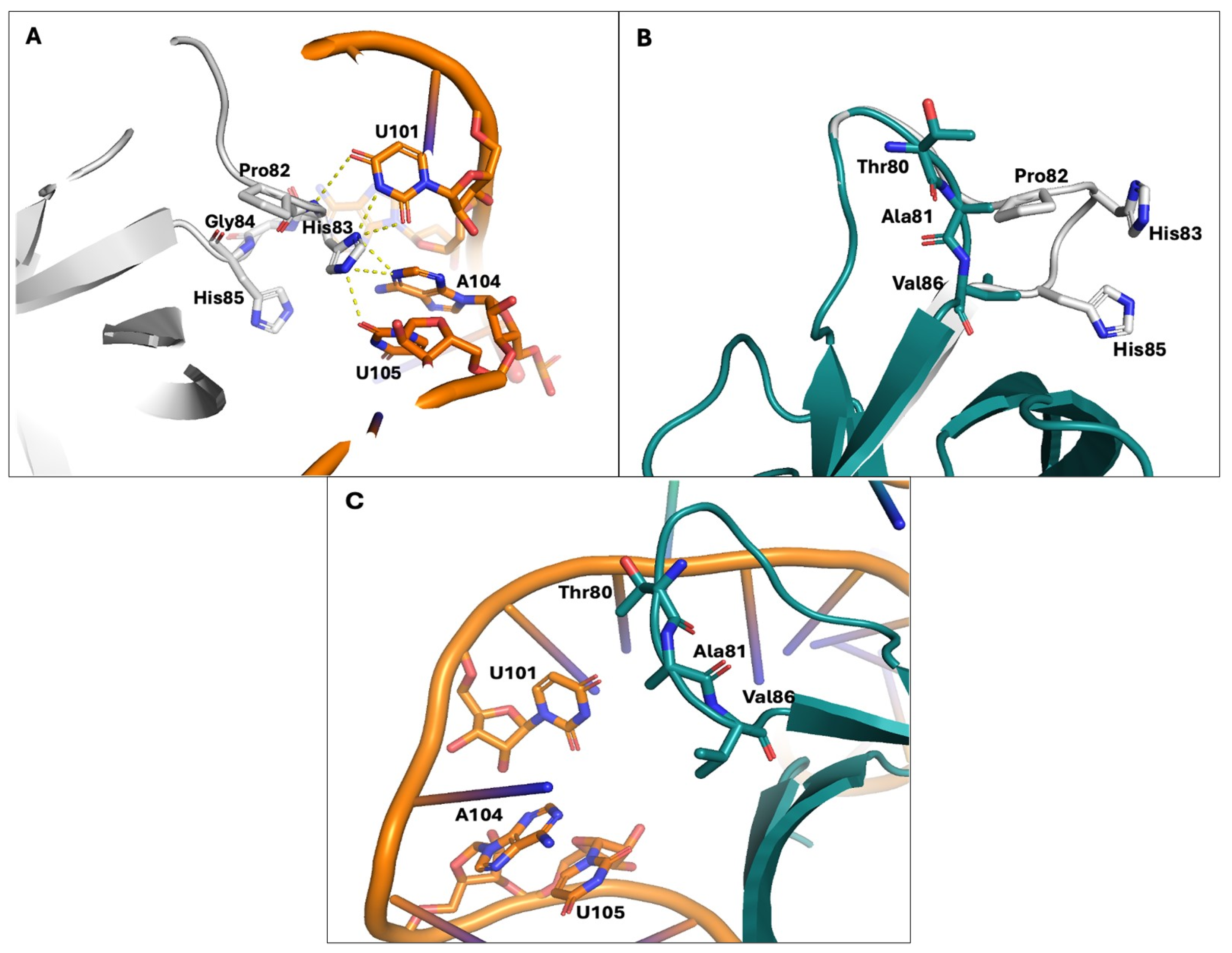
3.6. The 82–85 Nsp1 Domain Alters Viral mRNAs’ Binding Affinity
4. Discussion
5. Conclusions
6. Limitation of the Study
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- World Health Organization (WHO). COVID-19 Dashboard. Available online: https://data.who.int/dashboards/covid19/deaths?n=c (accessed on 31 December 2019).
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.K.; Blanco, M.R.; Bruce, E.A.; Honson, D.D.; Chen, L.M.; Chow, A.; Bhat, P.; Ollikainen, N.; Quinodoz, S.A.; Loney, C.; et al. SARS-CoV-2 Disrupts Splicing, Translation, and Protein Trafficking to Suppress Host Defenses. Cell 2020, 183, 1325–1339. [Google Scholar] [CrossRef] [PubMed]
- Shemesh, M.; Aktepe, T.E.; Deerain, J.M.; McAuley, J.L.; Audsley, M.D.; David, C.T.; Purcell, D.F.J.; Urin, V.; Hartmann, R.; Moseley, G.W.; et al. SARS-CoV-2 suppresses IFNβ production mediated by NSP1, 5, 6, 15, ORF6 and ORF7b but does not suppress the effects of added interferon. PLoS Pathog. 2021, 17, e1009800. [Google Scholar]
- Vazquez, C.; Swanson, S.E.; Negatu, S.G.; Dittmar, M.; Miller, J.; Ramage, H.R.; Cherry, S.; Jurado, K.A. SARS-CoV-2 viral proteins NSP1 and NSP13 inhibit interferon activation through distinct mechanisms. PLoS ONE 2021, 16, e0253089. [Google Scholar] [CrossRef] [PubMed]
- Gori Savellini, G.; Anichini, G.; Gandolfo, C.; Cusi, M.G. Nucleopore Traffic Is Hindered by SARS-CoV-2 ORF6 Protein to Efficiently Suppress IFN-β and IL-6 Secretion. Viruses 2022, 14, 1273. [Google Scholar] [CrossRef] [PubMed]
- Schubert, K.; Karousis, E.D.; Jomaa, A.; Scaiola, A.; Echeverria, B.; Gurzeler, L.A.; Leibundgut, M.; Thiel, V.; Mühlemann, O.; Ban, N. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020, 27, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Hu, Z.; Zhang, X.; Chen, M.; Wang, Z.; Xu, G.; Bi, Y.; Tong, Q.; Wang, M.; Sun, H.; et al. An R195K Mutation in the PA-X Protein Increases the Virulence and Transmission of Influenza A Virus in Mammalian Hosts. J. Virol. 2020, 94, e01817-19. [Google Scholar] [CrossRef]
- Zhang, R.; Li, Y.; Cowley, T.J.; Steinbrenner, A.D.; Phillips, J.M.; Yount, B.L.; Baric, R.S.; Weiss, S.R. The nsp1, nsp13, and M proteins contribute to the hepatotropism of murine coronavirus JHM.WU. J. Virol. 2015, 89, 3598–3609. [Google Scholar] [CrossRef] [PubMed]
- Züst, R.; Cervantes-Barragán, L.; Kuri, T.; Blakqori, G.; Weber, F.; Ludewig, B.; Thiel, V. Coronavirus non-structural protein 1 is a major pathogenicity factor: Implications for the rational design of coronavirus vaccines. PLoS Pathog. 2007, 3, e109. [Google Scholar] [CrossRef]
- Mendez, A.S.; Ly, M.; González-Sánchez, A.M.; Hartenian, E.; Ingolia, N.T.; Cate, J.H.; Glaunsinger, B.A. The N-terminal domain of SARS-CoV-2 nsp1 plays key roles in suppression of cellular gene expression and preservation of viral gene expression. Cell Rep. 2021, 37, 109841. [Google Scholar] [CrossRef]
- Gaucherand, L.; Porter, B.K.; Levene, R.E.; Price, E.L.; Schmaling, S.K.; Rycroft, C.H.; Kevorkian, Y.; McCormick, C.; Khaperskyy, D.A.; Gaglia, M.M. The Influenza A Virus Endoribonuclease PA-X Usurps Host mRNA Processing Machinery to Limit Host Gene Expression. Cell Rep. 2019, 27, 776–792. [Google Scholar] [CrossRef] [PubMed]
- Thoms, M.; Buschauer, R.; Ameismeier, M.; Koepke, L.; Denk, T.; Hirschenberger, M.; Kratzat, H.; Hayn, M.; Mackens-Kiani, T.; Cheng, J.; et al. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science 2020, 369, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Tidu, A.; Janvier, A.; Schaeffer, L.; Sosnowski, P.; Kuhn, L.; Hammann, P.; Westhof, E.; Eriani, G.; Martin, F. The viral protein NSP1 acts as a ribosome gatekeeper for shutting down host translation and fostering SARS-CoV-2 translation. RNA 2020, 27, 253–264. [Google Scholar] [CrossRef]
- Yuan, S.; Peng, L.; Park, J.J.; Hu, Y.; Devarkar, S.C.; Dong, M.B.; Shen, Q.; Wu, S.; Chen, S.; Lomakin, I.B.; et al. Nonstructural Protein 1 of SARS-CoV-2 Is a Potent Pathogenicity Factor Redirecting Host Protein Synthesis Machinery toward Viral RNA. Mol. Cell 2020, 80, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Malik, Y.A. Properties of Coronavirus and SARS-CoV-2. Malays. J. Pathol. 2020, 42, 3–11. [Google Scholar] [PubMed]
- Yao, H.; Song, Y.; Chen, Y.; Wu, N.; Xu, J.; Sun, C.; Zhang, J.; Weng, T.; Zhang, Z.; Wu, Z.; et al. Molecular Architecture of the SARS-CoV-2. Virus Cell 2020, 183, 730–738. [Google Scholar]
- Bai, C.; Zhong, Q.; Gao, G.F. Overview of SARS-CoV-2 genome-encoded proteins. Sci. China Life Sci. 2022, 65, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Arya, R.; Kumari, S.; Pandey, B.; Mistry, H.; Bihani, S.C.; Das, A.; Prashar, V.; Gupta, G.D.; Panicker, L.; Kumar, M. Structural insights into SARS-CoV-2 proteins. J. Mol. Biol. 2021, 433, 166725. [Google Scholar] [CrossRef] [PubMed]
- Redondo, N.; Zaldívar-López, S.; Garrido, J.J.; Montoya, M. SARS-CoV-2 Accessory Proteins in Viral Pathogenesis: Knowns and Unknowns. Front. Immunol. 2021, 12, 708264. [Google Scholar] [CrossRef]
- Gorkhali, R.; Koirala, P.; Rijal, S.; Mainali, A.; Baral, A.; Bhattarai, H.K. Structure and Function of Major SARS-CoV-2 and SARS-CoV Proteins. Bioinform. Biol. Insights 2021, 15, 11779322211025876. [Google Scholar] [CrossRef]
- Zandi, M.; Shafaati, M.; Kalantar-Neyestanaki, D.; Pourghadamyari, H.; Fani, M.; Soltani, S.; Kaleji, H.; Abbasi, S. The role of SARS-CoV-2 accessory proteins in immune evasion. Biomed. Pharmacother. 2022, 156, 113889. [Google Scholar] [CrossRef]
- Rashid, F.; Xie, Z.; Suleman, M.; Shah, A.; Khan, S.; Luo, S. Roles and functions of SARS-CoV-2 proteins in host immune evasion. Front. Immunol. 2022, 13, 940756. [Google Scholar] [CrossRef]
- Miorin, L.; Kehrer, T.; Sanchez-Aparicio, M.T.; Zhang, K.; Cohen, P.; Patel, R.S.; Cupic, A.; Makio, T.; Mei, M.; Moreno, E.; et al. SARS-CoV-2 Orf6 hijacks Nup98 to block STAT nuclear import and antagonize interferon signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 28344–28354. [Google Scholar] [CrossRef]
- Kamitani, W.; Narayanan, K.; Huang, C.; Lokugamage, K.; Ikegami, T.; Ito, N.; Kubo, H.; Makino, S. Severe acute respiratory syndrome coronavirus nsp1 protein suppresses host gene expression by promoting host mRNA degradation. Proc. Natl. Acad Sci. USA 2006, 103, 12885–12890. [Google Scholar] [CrossRef] [PubMed]
- Lokugamage, K.G.; Narayanan, K.; Huang, C.; Makino, S. Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation. J. Virol. 2012, 86, 13598–13608. [Google Scholar] [CrossRef]
- Schroeder, S.; Pott, F.; Niemeyer, D.; Veith, T.; Richter, A.; Muth, D.; Goffinet, C.; Müller, M.A.; Drosten, C. Interferon antagonism by SARS-CoV-2: A functional study using reverse genetics. Lancet Microbe 2021, 2, e210–e218. [Google Scholar] [CrossRef] [PubMed]
- Gori Savellini, G.; Anichini, G.; Cusi, M.G. SARS-CoV-2 omicron sub-lineages differentially modulate interferon response in human lung epithelial cells. Virus Res. 2023, 332, 199134. [Google Scholar] [CrossRef] [PubMed]
- Thorne, L.G.; Bouhaddou, M.; Reuschl, A.K.; Zuliani-Alvarez, L.; Polacco, B.; Pelin, A.; Batra, J.; Whelan, M.V.X.; Hosmillo, M.; Fossati, A.; et al. Evolution of enhanced innate immune evasion by SARS-CoV-2. Nature 2022, 602, 487–495. [Google Scholar] [CrossRef]
- Zheng, Y.; Deng, J.; Han, L.; Zhuang, M.W.; Xu, Y.; Zhang, J.; Nan, M.L.; Xiao, Y.; Zhan, P.; Liu, X.; et al. SARS-CoV-2 NSP5 and N protein counteract the RIG-I signaling pathway by suppressing the formation of stress granules. Signal Transduct. Target. Ther. 2022, 7, 22. [Google Scholar] [CrossRef]
- Addetia, A.; Lieberman, N.A.P.; Phung, Q.; Hsiang, T.Y.; Xie, H.; Roychoudhury, P.; Shrestha, L.; Loprieno, M.A.; Huang, M.L.; Gale, M., Jr.; et al. SARS-CoV-2 ORF6 Disrupts Bidirectional Nucleocytoplasmic Transport through Interactions with Rae1 and Nup98. mBio 2021, 12, e00065-21. [Google Scholar] [CrossRef]
- Brant, A.C.; Tian, W.; Majerciak, V.; Yang, W.; Zheng, Z.M. SARS-CoV-2: From its discovery to genome structure, transcription, and replication. Cell Biosci. 2021, 11, 136. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi-Dehcheshmeh, M.; Moghbeli, S.M.; Rahimirad, S.; Alanazi, I.O.; Shehri, Z.S.A.; Ebrahimie, E. Transcription Regulatory Sequence in the 5′ Untranslated Region of SARS-CoV-2 Is Vital for Virus Replication with an Altered Evolutionary Pattern against Human Inhibitory MicroRNAs. Cells 2021, 10, 319. [Google Scholar] [CrossRef]
- Shi, M.; Wang, L.; Fontana, P.; Vora, S.; Zhang, Y.; Fu, T.M.; Lieberman, J.; Wu, H. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. bioRxiv 2020. [Google Scholar] [CrossRef]
- Long, S. SARS-CoV-2 Subgenomic RNAs: Characterization, Utility, and Perspectives. Viruses 2021, 13, 1923. [Google Scholar] [CrossRef]
- Mori, A.; Lavezzari, D.; Pomari, E.; Deiana, M.; Piubelli, C.; Capobianchi, M.R.; Castilletti, C. sgRNAs: A SARS-CoV-2 emerging issue. Asp. Mol. Med. 2023, 1, 100008. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Lokugamage, K.G.; Rozovics, J.M.; Narayanan, K.; Semler, B.L.; Makino, S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: Viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011, 7, e1002433. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Huang, C.; Lokugamage, K.; Kamitani, W.; Ikegami, T.; Tseng, C.T.; Makino, S. Severe acute respiratory syndrome coronavirus nsp1 suppresses host gene expression, including that of type I interferon, in infected cells. J. Virol. 2008, 82, 4471–4479. [Google Scholar] [CrossRef]
- Lapointe, C.P.; Grosely, R.; Johnson, A.G.; Wang, J.; Fernández, I.S.; Puglisi, J.D. Dynamic competition between SARS-CoV-2 NSP1 and mRNA on the human ribosome inhibits translation initiation. Proc. Natl. Acad. Sci. USA 2021, 118, e2017715118. [Google Scholar] [CrossRef]
- Levene, R.E.; Shrestha, S.D.; Gaglia, M.M. The influenza A virus host shutoff factor PA-X is rapidly turned over in a strain-specific manner. J. Virol. 2021, 95, e02312-20. [Google Scholar] [CrossRef]
- Sarnow, P.; Cevallos, R.C.; Jan, E. Takeover of host ribosomes by divergent IRES elements. Biochem. Soc. Trans. 2005, 33, 1479–1482. [Google Scholar] [CrossRef]
- Lin, J.W.; Tang, C.; Wei, H.C.; Du, B.; Chen, C.; Wang, M.; Zhou, Y.; Yu, M.X.; Cheng, L.; Kuivanen, S.; et al. Genomic monitoring of SARS-CoV-2 uncovers an Nsp1 deletion variant that modulates type I interferon response. Cell Host Microbe 2021, 29, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Tsueng, G.; Mullen, J.L.; Alkuzweny, M.; Cano, M.; Rush, B.; Haag, E.; Lin, J.; Welzel, D.J.; Zhou, X.; Qian, Z.; et al. Outbreak.info Research Library: A standardized, searchable platform to discover and explore COVID-19 resources. Nat. Methods 2023, 20, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Gaglia, M.M.; Covarrubias, S.; Wong, W.; Glaunsinger, B.A. A common strategy for host RNA degradation by divergent viruses. J. Virol. 2012, 86, 9527–9530. [Google Scholar] [CrossRef] [PubMed]
- Finkel, Y.; Mizrahi, O.; Nachshon, A.; Weingarten-Gabbay, S.; Morgenstern, D.; Yahalom-Ronen, Y.; Tamir, H.; Achdout, H.; Stein, D.; Israeli, O.; et al. The coding capacity of SARS-CoV-2. Nature 2021, 589, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, K.; Ramirez, S.I.; Lokugamage, K.G.; Makino, S. Coronavirus nonstructural protein 1: Common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015, 202, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Vora, S.M.; Fontana, P.; Mao, T.; Leger, V.; Zhang, Y.; Fu, T.M.; Lieberman, J.; Gehrke, L.; Shi, M.; Wang, L.; et al. Targeting stem-loop 1 of the SARS-CoV-2 5′ UTR to suppress viral translation and Nsp1 evasion. Proc. Natl. Acad. Sci. USA 2022, 119, e2117198119. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kamitani, W.; DeDiego, M.L.; Enjuanes, L.; Matsuura, Y. Severe acute respiratory syndrome coronavirus nsp1 facilitates efficient propagation in cells through a specific translational shutoff of host mRNA. J. Virol. 2012, 86, 11128–11137. [Google Scholar] [CrossRef] [PubMed]
- Sakuraba, S.; Xie, Q.; Kasahara, K.; Iwakiri, J.; Kono, H. Extended ensemble simulations of a SARS-CoV-2 nsp1-5′-UTR complex. PLoS Comput. Biol. 2022, 18, e1009804. [Google Scholar] [CrossRef]
- Afsar, M.; Narayan, R.; Akhtar, M.N.; Das, D.; Rahil, H.; Nagaraj, S.K.; Eswarappa, S.M.; Tripathi, S.; Hussain, T. Drug targeting Nsp1-ribosomal complex shows antiviral activity against SARS-CoV-2. eLife 2022, 11, e74877. [Google Scholar] [CrossRef]
- Khan, A.R.; Misdary, C.; Yegya-Raman, N.; Kim, S.; Narayanan, N.; Siddiqui, S.; Salgame, P.; Radbel, J.; Groote, F.; Michel, C.; et al. Montelukast in hospitalized patients diagnosed with COVID-19. J. Asthma 2022, 59, 780–786. [Google Scholar] [CrossRef]
- Vankadari, N.; Jeyasankar, N.N.; Lopes, W.J. Structure of the SARS-CoV-2 Nsp1/5′-untranslated region complex and implications for potential therapeutic targets, a vaccine, and virulence. J. Phys. Chem. Lett. 2020, 11, 9659–9668. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Zheng, Y.; Zeng, X.; He, B.; Cheng, W. Structural biology of SARS-CoV-2: Open the door for novel therapies. Signal Transduct. Target. Ther. 2022, 7, 26. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gori Savellini, G.; Anichini, G.; Manetti, F.; Trivisani, C.I.; Cusi, M.G. Deletion of 82–85 N-Terminal Residues in SARS-CoV-2 Nsp1 Restricts Virus Replication. Viruses 2024, 16, 689. https://doi.org/10.3390/v16050689
Gori Savellini G, Anichini G, Manetti F, Trivisani CI, Cusi MG. Deletion of 82–85 N-Terminal Residues in SARS-CoV-2 Nsp1 Restricts Virus Replication. Viruses. 2024; 16(5):689. https://doi.org/10.3390/v16050689
Chicago/Turabian StyleGori Savellini, Gianni, Gabriele Anichini, Fabrizio Manetti, Claudia Immacolata Trivisani, and Maria Grazia Cusi. 2024. "Deletion of 82–85 N-Terminal Residues in SARS-CoV-2 Nsp1 Restricts Virus Replication" Viruses 16, no. 5: 689. https://doi.org/10.3390/v16050689
APA StyleGori Savellini, G., Anichini, G., Manetti, F., Trivisani, C. I., & Cusi, M. G. (2024). Deletion of 82–85 N-Terminal Residues in SARS-CoV-2 Nsp1 Restricts Virus Replication. Viruses, 16(5), 689. https://doi.org/10.3390/v16050689







