Neuraminidase Inhibitor Susceptibility Testing in Human Influenza Viruses: A Laboratory Surveillance Perspective
Abstract
:1. Introduction
2. Results and Discussion
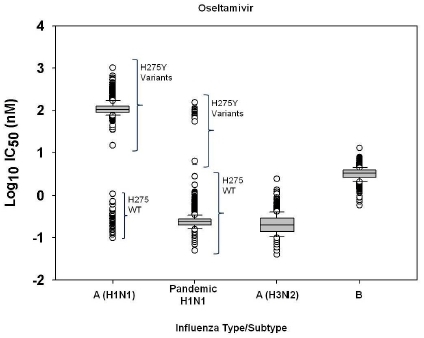
2.1. Susceptibility of seasonal influenza virus isolates to neuraminidase inhibitors
2.2. Susceptibility of 2009 pandemic H1N1 viruses to neuraminidase inhibitors
2.3. Challenges of defining neuraminidase inhibitor resistance for surveillance
3. Experimental Section
3.1. Viruses and cells
3.2. NA inhibitors
3.3. NA inhibition assays
3.4. Statistical analysis
3.5. Pyrosequencing
3.6. Sequencing by dideoxy chain termination method
4. Conclusions
Acknowledgments
Disclaimer
References and Notes
- Moscona, A. Medical management of influenza infection. Annu. Rev. Med. 2008, 59, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Bright, R.A.; Medina, M.J.; Xu, X.; Perez-Oronoz, G.; Wallis, T.R.; Davis, X.M.; Povinelli, L.; Cox, N.J.; Klimov, A.I. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet. 2005, 366, 1175–1181. [Google Scholar] [CrossRef] [PubMed]
- Deyde, V.M.; Xu, X.; Bright, R.A.; Shaw, M.; Smith, C.B.; Zhang, Y.; Shu, Y.; Gubareva, L.V.; Cox, N.J.; Klimov, A.I. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J. Infect. Dis. 2007, 196, 249–257. [Google Scholar] [CrossRef] [PubMed]
- CDC Website. http://www.cdc.gov/h1n1flu/eua/peramivir.htm. (accessed on 28 May 2010).
- Shionogi and Company Website. http://www.shionogi.co.jp/ir_en/news/detail/e_100126.pdf. (accessed on 28 May 2010).
- Colman, P.M.; Hoyne, P.A.; Lawrence, M.C. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J. Virol. 1993, 67, 2972–2980. [Google Scholar] [CrossRef] [PubMed]
- Ferraris, O.; Lina, B. Mutations of neuraminidase implicated in neuraminidase inhibitors resistance. J. Clin. Virol. 2008, 41, 13–19. [Google Scholar] [CrossRef]
- Monto, A.S.; McKimm-Breschkin, J.L.; Macken, C.; Hampson, A.W.; Hay, A.; Klimov, A.; Tashiro, M.; Webster, R.G.; Aymard, M.; Hayden, F.G.; Zambon, M. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob. Agents Chemother. 2006, 50, 2395–2402. [Google Scholar] [CrossRef]
- Mungall, B.A.; Xu, X.; Klimov, A. Surveillance of influenza isolates for susceptibility to neuraminidase inhibitors during the 2000-2002 influenza seasons. Virus Res. 2004, 103, 195–197. [Google Scholar] [CrossRef]
- Sheu, T.G.; Deyde, V.M.; Okomo-Adhiambo, M.; Garten, R.J.; Xu, X.; Bright, R.A.; Butler, E.N.; Wallis, T.R.; Klimov, A.I.; Gubareva, L.V. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob. Agents Chemother. 2008, 52, 3284–3292. [Google Scholar] [CrossRef]
- Hauge, S.H.; Dudman, S.; Borgen, K.; Lackenby, A.; Hungnes, O. Oseltamivir-resistant influenza viruses A (H1N1), Norway, 2007-08. Emerg. Infect. Dis. 2009, 15, 155–162. [Google Scholar] [CrossRef]
- CDC. Influenza activity - United States and worldwide, 2007-08 season. Morb. Mortal. Wkly. Rep. 2008, 57, 692–697. [Google Scholar]
- Lackenby, A.; Hungnes, O.; Dudman, S.G.; Meijer, A.; Paget, W.J.; Hay, A.J.; Zambon, M.C. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro. Surveill. 2008, 13, 8026. [Google Scholar] [CrossRef] [PubMed]
- Meijer, A.; Lackenby, A.; Hungnes, O.; Lina, B.; van der Werf, S.; Schweiger, B.; Opp, M.; Paget, J.; van de Kassteele, J.; Hay, A.; Zambon, M. Oseltamivir-resistant influenza virus A (H1N1), Europe, 2007-08 Season. Emerg. Infect. Dis. 2009, 15, 552–560. [Google Scholar] [CrossRef] [PubMed]
- Besselaar, T.G.; Naidoo, D.; Buys, A.; Gregory, V.; McAnerney, J.; Manamela, J.M.; Blumberg, L.; Schoub, B.D. Widespread oseltamivir resistance in influenza A viruses (H1N1), South Africa. Emerg. Infect. Dis. 2008, 14, 1809–1810. [Google Scholar] [CrossRef] [PubMed]
- CDC. Update: influenza activity - United States, September 28, 2008-January 31, 2009. Morb. Mortal. Wkly. Rep. 2009, 58, 115–119. [Google Scholar]
- CDC. Update: influenza activity - United States, September 28, 2008-April 4, 2009, and composition of the 2009-10 influenza vaccine. Morb. Mortal. Wkly. Rep. 2009, 58, 369–374. [Google Scholar]
- Baranovich, T.; Saito, R.; Suzuki, Y.; Zaraket, H.; Dapat, C.; Caperig-Dapat, I.; Oguma, T.; Shabana, I.I.; Saito, T.; Suzuki, H. Emergence of H274Y oseltamivir-resistant A(H1N1) influenza viruses in Japan during the 2008-2009 season. J. Clin. Virol. 2010, 47, 23–28. [Google Scholar] [CrossRef] [PubMed]
- Mungall, B.A.; Xu, X.; Klimov, A. Assaying susceptibility of avian and other influenza A viruses to zanamivir: comparison of fluorescent and chemiluminescent neuraminidase assays. Avian Dis. 2003, 47, 1141–1144. [Google Scholar] [CrossRef]
- Wetherall, N.T.; Trivedi, T.; Zeller, J.; Hodges-Savola, C.; McKimm-Breschkin, J.L.; Zambon, M.; Hayden, F.G. Evaluation of neuraminidase enzyme assays using different substrates to measure susceptibility of influenza virus clinical isolates to neuraminidase inhibitors: report of the neuraminidase inhibitor susceptibility network. J. Clin. Microbiol. 2003, 41, 742–750. [Google Scholar] [CrossRef]
- McKimm-Breschkin, J.; Trivedi, T.; Hampson, A.; Hay, A.; Klimov, A.; Tashiro, M.; Hayden, F.; Zambon, M. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob. Agents Chemother. 2003, 47, 2264–2272. [Google Scholar] [CrossRef]
- Mishin, V.P.; Hayden, F.G.; Gubareva, L.V. Susceptibilities of antiviral-resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob. Agents Chemother. 2005, 49, 4515–4520. [Google Scholar] [CrossRef]
- Buxton, R.C.; Edwards, B.; Juo, R.R.; Voyta, J.C.; Tisdale, M.; Bethell, R.C. Development of a sensitive chemiluminescent neuraminidase assay for the determination of influenza virus susceptibility to zanamivir. Anal. Biochem. 2000, 280, 291–300. [Google Scholar] [CrossRef]
- Potier, M.; Mameli, L.; Belisle, M.; Dallaire, L.; Melancon, S.B. Fluorometric assay of neuraminidase with a sodium (4-methylumbelliferyl-alpha-D-N-acetylneuraminate) substrate. Anal. Biochem. 1979, 94, 287–296. [Google Scholar] [CrossRef]
- Hurt, A.C.; Holien, J.K.; Parker, M.; Kelso, A.; Barr, I.G. Zanamivir-Resistant Influenza Viruses with a Novel Neuraminidase Mutation. J. Virol. 2009, 83, 10366-73. [Google Scholar] [CrossRef]
- Okomo-Adhiambo, M.; Nguyen, H.T.; Sleeman, K.; Sheu, T.G.; Deyde, V.M.; Garten, R.J.; Xu, X.; Shaw, M.W.; Klimov, A.I.; Gubareva, L.V. Host cell selection of influenza neuraminidase variants: implications for drug resistance monitoring in A(H1N1) viruses. Antiviral Res. 2010, 85, 381–388. [Google Scholar] [CrossRef]
- NISN Website. http://www.nisn.org/v_analysis_guidance_ic50.php. (accessed on 28 May 2010).
- Tisdale, M. Monitoring of viral susceptibility: new challenges with the development of influenza NA inhibitors. Rev. Med. Virol. 2000, 10, 45–55. [Google Scholar] [CrossRef]
- Deyde, V.M.; Okomo-Adhiambo, M.; Sheu, T.G.; Wallis, T.R.; Fry, A.; Dharan, N.; Klimov, A.I.; Gubareva, L.V. Pyrosequencing as a tool to detect molecular markers of resistance to neuraminidase inhibitors in seasonal influenza A viruses. Antiviral Res. 2009, 81, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Deyde, V.M.; Sheu, T.G.; Trujillo, A.A.; Okomo-Adhiambo, M.; Garten, R.; Klimov, A.I.; Gubareva, L.V. Detection of molecular markers of drug resistance in 2009 pandemic influenza A (H1N1) viruses by pyrosequencing. Antimicrob. Agents Chemother. 2010, 54, 1102–1110. [Google Scholar] [CrossRef]
- Chidlow, G.R.; Harnett, G.B.; Williams, S.H.; Tempone, S.S.; Speers, D.J.; Hurt, A.C.; Deng, Y. M.; Smith, D.W. The detection of oseltamivir-resistant pandemic influenza A/H1N1 2009 viruses using a real-time RT-PCR assay. J. Virol. Methods. 2010, 169, 47–51. [Google Scholar] [CrossRef]
- van der Vries, E.; Jonges, M.; Herfst, S.; Maaskant, J.; van der Linden, A.; Guldemeester, J.; Aron, G. I.; Bestebroer, T.M.; Koopmans, M.; Meijer, A.; Fouchier, R.A.; Osterhaus, A.D.; Boucher, C.A.; Schutten, M. Evaluation of a rapid molecular algorithm for detection of pandemic influenza A (H1N1) 2009 virus and screening for a key oseltamivir resistance (H275Y) substitution in neuraminidase. J. Clin. Virol. 2010, 47, 34–37. [Google Scholar] [CrossRef] [PubMed]
- CDC Update: swine influenza A (H1N1) infections--California and Texas, April 2009. Morb. Mortal. Wkly. Rep. 2009, 58, 435–437.
- Tashiro, M.; McKimm-Breschkin, J.L.; Saito, T.; Klimov, A.; Macken, C.; Zambon, M.; Hayden, F.G. Surveillance for neuraminidase-inhibitor-resistant influenza viruses in Japan, 1996-2007. Antivir. Ther. 2009, 14, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Hurt, A.C.; Barr, I.G.; Hartel, G.; Hampson, A.W. Susceptibility of human influenza viruses from Australasia and South East Asia to the neuraminidase inhibitors zanamivir and oseltamivir. Antiviral Res. 2004, 62, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Jonges, M.; van der Lubben, I.M.; Dijkstra, F.; Verhoef, L.; Koopmans, M.; Meijer, A. Dynamics of antiviral-resistant influenza viruses in the Netherlands, 2005-2008. Antiviral Res. 2009, 83, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.P.; Gregory, V.; Collins, P.; Kloess, J.; Wharton, S.; Cattle, N.; Lackenby, A.; Daniels, R.; Hay, A. Neuraminidase Receptor Binding Variants of Human Influenza A(H3N2) Viruses due to Substitution of Aspartic acid 151 in the Catalytic Site - Role in Virus Attachment? J. Virol. 2010, 84, 6769-81. [Google Scholar] [CrossRef] [PubMed]
- Hurt, A.C.; Iannello, P.; Jachno, K.; Komadina, N.; Hampson, A.W.; Barr, I.G.; McKimm-Breschkin, J.L. Neuraminidase inhibitor-resistant and -sensitive influenza B viruses isolated from an untreated human patient. Antimicrob. Agents Chemother. 2006, 50, 1872–1874. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Sheu, T.G.; Mishin, V.P.; Klimov, A.I.; Gubareva, L.V. Assessment of Pandemic and Seasonal Influenza A (H1N1) Virus Susceptibility to Neuraminidase Inhibitors in Three Enzyme Activity Inhibition Assays. Antimicrob. Agents Chemother. 2010, 54, 3671–3677. [Google Scholar] [CrossRef]
- Sugaya, N.; Mitamura, K.; Yamazaki, M.; Tamura, D.; Ichikawa, M.; Kimura, K.; Kawakami, C.; Kiso, M.; Ito, M.; Hatakeyama, S.; Kawaoka, Y. Lower clinical effectiveness of oseltamivir against influenza B contrasted with influenza A infection in children. Clin. Infect. Dis. 2007, 44, 197–202. [Google Scholar] [CrossRef]
- Sato, M.; Saito, R.; Sato, I.; Tanabe, N.; Shobugawa, Y.; Sasaki, A.; Li, D.; Suzuki, Y.; Sato, M.; Sakai, T.; Oguma, T.; Tsukada, H.; Gejyo, F.; Suzuki, H. Effectiveness of oseltamivir treatment among children with influenza A or B virus infections during four successive winters in Niigata City, Japan. Tohoku J. Exp. Med. 2008, 214, 113–120. [Google Scholar] [CrossRef]
- Ferraris, O.; Kessler, N.; Lina, B. Sensitivity of influenza viruses to zanamivir and oseltamivir: a study performed on viruses circulating in France prior to the introduction of neuraminidase inhibitors in clinical practice. Antiviral Res. 2005, 68, 43–48. [Google Scholar] [CrossRef]




| Strain Designation | Type/Subtype | NA Mutations (Genotype) | Oseltamivir Susceptibility | Mean IC50a ± SDb, nM (Fold Difference c) | ||
|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | Peramivir | ||||
| A/Washington/10/2008 | A (H1N1); Seasonal | WTd | Se | 0.23±0.07 (1) | 0.24±0.08 (1) | 0.09±0.02 (1) |
| A/Florida/21/2008 | A (H1N1); Seasonal | H275Y | Rf | 94.82±27.04 (386) | 0.31±0.08 (1) | 11.27±1.36 (134) |
| A/Washington/01/2007 | A (H3N2); Seasonal | WT | S | 0.15±0.00 (1) | 0.47±0.04 (1) | 0.12±0.02 (1) |
| A/Texas/12/2007 | A (H3N2); Seasonal | E119V | R | 4.19±0.33 (30) | 0.51±0.05 (1) | 0.14±0.02 (1) |
| B/Memphis/20/1996 | B; Seasonal | WT | S | 1.61±0.21 (1) | 1.94±0.21 (1) | 0.31±0.06 (1) |
| B/Memphis/20/1996 | B; Seasonal | R152K | R | 82.62±2.96 (58) | 26.35±13.59 (22) | 52.80±3.51 (176) |
| A/California/07/2009 | H1N1; Pandemic | WT | S | 0.21±0.03 (1) | 0.26±0.04 (1) | 0.06±0.02 (1) |
| A/Texas/48/2009 | H1N1; Pandemic | H275Y | R | 79.94±0.06 (348) | 0.36±0.07 (1) | 10.06±0.01 (164) |
| Subtype | Isolates analyzed (n) | Mean IC50 a (±SD)b, nM | ||||||
|---|---|---|---|---|---|---|---|---|
| Oseltamivir | Zanamivir | |||||||
| JASPR | Robosage | rc | JASPR | Robosage | r | |||
| Seasonal A/H1N1 (H275 wildtype)d | 8 | 0.22±0.05 | 0.26±0.07 | 0.970 | 0.47±0.34 | 0.56±0.47 | 0.999 | |
| Seasonal A/H1N1 (H275Y variants)e | 106 | 113.38±45.63 | 128.57±55.93 | 0.986 | 0.50±0.23 | 0.58±0.30 | 0.990 | |
| Pandemic H1N1 | 100 | 0.21±0.06 | 0.25±0.06 | 0.939 | 0.28±0.10 | 0.31±0.13 | 0.988 | |
| A/H3N2 | 125 | 0.24±0.13 | 0.27±0.16 | 0.994 | 1.30±0.93 | 1.48±1.08 | 0.994 | |
| B | 106 | 3.56±0.94 | 3.84±1.05 | 0.990 | 3.93±1.32 | 4.19±1.45 | 0.995 | |
| NAI | Influenza Type/Subtype | Isolates analyzed (n)* | IC50 (nM)a | |||||
|---|---|---|---|---|---|---|---|---|
| Range | Mean (±SD)b | Median | IQRd | X0.75 e | Statistical Cutoff f | |||
| Oseltamivir | A(H1N1) (H275 wildtype)g | 98 | 0.10-.0.49 | 0.23±0.08 | 0.21 | 0.10 | 0.28 | 0.58 |
| A(H1N1) (H275Y variants)h | 1430 | 34.69-1023.68 | 123.32±65.90 | 105.13 | -- | -- | -- | |
| A/H3N2 | 833 | 0.04-1.38 | 0.24±0.15 | 0.20 | 0.15 | 0.29 | 0.74 | |
| B | 913 | 0.59-7.75 | 3.41±0.99 | 3.38 | 1.27 | 4.01 | 7.82 | |
| Zanamivir | A(H1N1) (H275 wildtype)g | 102 | 0.13-1.03 | 0.35±0.15 | 0.31 | 0.17 | 0.54 | 1.05 |
| A(H1N1) (H275Y variants)h | 1424 | 0.07-3.49 | 0.51±0.27 | 0.45 | -- | -- | -- | |
| A(H3N2) | 812 | 0.22-4.24 | 1.23±0.80 | 0.97 | 0.91 | 1.58 | 4.31 | |
| B | 911 | 0.27-8.77 | 3.34±1.31 | 3.27 | 1.94 | 4.24 | 10.06 | |
| Peramivir | A(H1N1) (H275 wildtype) | 19 | 0.05-0.17 | 0.10±0.03 | 0.10 | 0.05 | 0.13 | 0.28 |
| A(H1N1) (H275Y variants) | 215 | 2.46-510.57 | 30.56±66.06 | 12.95 | -- | -- | -- | |
| A/H3N2 | 219 | 0.05-0.94 | 0.17±0.10 | 0.14 | 0.07 | 0.19 | 0.40 | |
| B | 52 | 0.24-1.10 | 0.56±0.18 | 0.56 | 0.23 | 0.68 | 1.37 | |
| NAI | Pandemic H1N1 virus isolates | Isolates analyzed (n)* | IC50 (nM)a | |||||
|---|---|---|---|---|---|---|---|---|
| Range | Mean (±SD)b | Median | IQRc | X0.75 d | Statistical Cutoff e | |||
| Oseltamivir | H275 wildtypef | 2243 | 0.05-1.78 | 0.25±0.11 | 0.24 | 0.08 | 0.28 | 0.52 |
| H275Y variantsg | 14 | 54.21-155.00 | 87.57±25.53 | 80.30 | -- | -- | -- | |
| Zanamivir | H275 wildtypef | 2233 | 0.08-1.03 | 0.31±0.08 | 0.30 | 0.11 | 0.36 | 0.69 |
| H275Y variantsg | 14 | 0.27-0.53 | 0.38±0.08 | 0.36 | ||||
| Peramivir | H275 wildtype | 538 | 0.03-0.35 | 0.08±0.04 | 0.07 | 0.04 | 0.10 | 0.22 |
| H275Y variants | 11 | 8.1-12.91 | 10.40±1.37 | 10.29 | -- | -- | -- | |
© 2010 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Okomo-Adhiambo, M.; Sleeman, K.; Ballenger, K.; Nguyen, H.T.; Mishin, V.P.; Sheu, T.G.; Smagala, J.; Li, Y.; Klimov, A.I.; Gubareva, L.V. Neuraminidase Inhibitor Susceptibility Testing in Human Influenza Viruses: A Laboratory Surveillance Perspective. Viruses 2010, 2, 2269-2289. https://doi.org/10.3390/v2102269
Okomo-Adhiambo M, Sleeman K, Ballenger K, Nguyen HT, Mishin VP, Sheu TG, Smagala J, Li Y, Klimov AI, Gubareva LV. Neuraminidase Inhibitor Susceptibility Testing in Human Influenza Viruses: A Laboratory Surveillance Perspective. Viruses. 2010; 2(10):2269-2289. https://doi.org/10.3390/v2102269
Chicago/Turabian StyleOkomo-Adhiambo, Margaret, Katrina Sleeman, Kristina Ballenger, Ha T. Nguyen, Vasiliy P. Mishin, Tiffany G. Sheu, James Smagala, Yan Li, Alexander I. Klimov, and Larisa V. Gubareva. 2010. "Neuraminidase Inhibitor Susceptibility Testing in Human Influenza Viruses: A Laboratory Surveillance Perspective" Viruses 2, no. 10: 2269-2289. https://doi.org/10.3390/v2102269