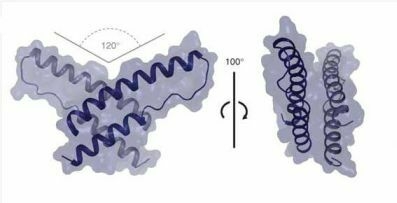
A Long-Awaited Structure Is Rev-ealed
Abstract
:Acknowledgements
References and Notes
- Daugherty, M.D.; D’Orso, I.; Frankel, A.D. A solution to limited genomic capacity: Using adaptable binding surfaces to assemble the functional HIV Rev oligomer on RNA. Mol. Cell 2008, 31, 824–834. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, M.D.; Booth, D.S.; Jayaraman, B.; Cheng, Y.; Frankel, A.D. HIV Rev response element (RRE) directs assembly of the Rev homooligomer into discrete asymmetric complexes. Proc. Nat. Acad. Sci. U. S. A. 2010, 107, 12481–12486. [Google Scholar] [CrossRef] [PubMed]
- Daugherty, M.D.; Liu, B.; Frankel, A.D. Structural basis for cooperative RNA binding and export complex assembly by HIV Rev. Nat. Struct. Mol. Biol. 2010, 17, 1337–1342. [Google Scholar] [CrossRef] [PubMed]
- DiMattia, M.A.; Watts, N.R.; Stahl, S.J.; Rader, C.; Wingfield, P.T.; Stuart, D.I.; Steven, A.C.; Grimes, J.M. Implications of the HIV-1 Rev dimer structure at 3.2 a resolution for multimeric binding to the Rev response element. Proc. Nat. Acad. Sci. U. S. A. 2010, 107, 5810–5814. [Google Scholar] [CrossRef]
- Feinberg, M.B.; Jarrett, R.F.; Aldovini, A.; Gallo, R.C.; Wong-Staal, F. HTLV-III expression and production involve complex regulation at the levels of splicing and translation of viral RNA. Cell 1986, 46, 807–817. [Google Scholar] [CrossRef]
- Sodroski, J.; Goh, W.C.; Rosen, C.; Dayton, A.; Terwilliger, E.; Haseltine, W. A second post-transcriptional trans-activator gene required for HTLV-III replication. Nature 1986, 321, 412–417. [Google Scholar] [CrossRef]
- Pollard, V.W.; Malim, M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998, 52, 491–532. [Google Scholar] [CrossRef]
- Daly, T.J.; Cook, K.S.; Gray, G.S.; Maione, T.E.; Rusche, J.R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 1989, 342, 816–819. [Google Scholar] [CrossRef]
- Zapp, M.L.; Green, M.R. Sequence-specific RNA binding by the HIV-1 Rev protein. Nature 1989, 342, 714–716. [Google Scholar] [CrossRef]
- Cochrane, A.W.; Chen, C.H.; Rosen, C.A. Specific interaction of the human immunodeficiency virus Rev protein with a structured region in the Env mRNA. Proc. Nat. Acad. Sci. U. S. A. 1990, 87, 1198–1202. [Google Scholar] [CrossRef]
- Daefler, S.; Klotman, M.E.; Wong, S.F. Trans-activating Rev protein of the human immunodeficiency virus 1 interacts directly and specifically with its target RNA. Proc. Nat. Acad. Sci. U. S. A. 1990, 87, 4571–4575. [Google Scholar] [CrossRef] [PubMed]
- Heaphy, S.; Dingwall, C.; Ernberg, I.; Gait, M.J.; Green, S.M.; Karn, J.; Lowe, A.D.; Singh, M.; Skinner, M.A. HIV-1 regulator of virion expression (Rev) protein binds to an RNA stem-loop structure located within the Rev response element region. Cell 1990, 60, 685–693. [Google Scholar] [CrossRef] [PubMed]
- Holland, S.M.; Ahmad, N.; Maitra, R.K.; Wingfield, P.; Venkatesan, S. Human immunodeficiency virus Rev protein recognizes a target sequence in Rev-responsive element RNA within the context of RNA secondary structure. J. Virol. 1990, 64, 5966–5975. [Google Scholar] [CrossRef] [PubMed]
- Malim, M.H.; Tiley, L.S.; McCarn, D.F.; Rusche, J.R.; Hauber, J.; Cullen, B.R. HIV-1 structural gene expression requires binding of the Rev trans-activator to its RNA target sequence. Cell 1990, 60, 675–683. [Google Scholar] [CrossRef]
- Rosen, C.A.; Terwilliger, E.; Dayton, A.; Sodroski, J.G.; Haseltine, W.A. Intragenic cis-acting Art gene-responsive sequences of the human immunodeficiency virus. Proc. Nat. Acad. Sci. U. S. A. 1988, 85, 2071–2075. [Google Scholar] [CrossRef]
- Hadzopoulou-Cladaras, M.; Felber, B.K.; Cladaras, C.; Athanassopoulos, A.; Tse, A.; Pavlakis, G.N. The Rev (Trs/Art) protein of human immunodeficiency virus type 1 affects viral mRNA and protein expression via a cis-acting sequence in the Env region. J. Virol. 1989, 63, 1265–1274. [Google Scholar] [CrossRef]
- Hammarskjöld, M.-L.; Heimer, J.; Hammarskjöld, B.; Sangwan, I.; Albert, L.; Rekosh, D. Regulation of human immunodeficiency virus env expression by the Rev gene product. J. Virol. 1989, 63, 1959–1966. [Google Scholar] [CrossRef]
- Malim, M.H.; Hauber, J.; Le, S.V.; Maizel, J.V.; Cullen, B.R. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 1989, 338, 254–257. [Google Scholar] [CrossRef]
- Felber, B.K.; Hadzopoulou-Cladaras, M.; Cladaras, C.; Copeland, T.; Pavlakis, G.N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Nat. Acad. Sci. U. S. A. 1989, 86, 1496–1499. [Google Scholar] [CrossRef]
- Legrain, P.; Seraphin, B.; Rosbash, M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol. Cell. Biol. 1988, 8, 3755–3760. [Google Scholar]
- Chang, D.D.; Sharp, P.A. Regulation by HIV Rev depends upon recognition of splice sites. Cell 1989, 59, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Hope, T.J.; Bond, B.L.; McDonald, D.; Klein, N.P.; Parslow, T.G. Effector domains of human immunodeficiency virus type 1 Rev and human T- cell leukemia virus type I Rex are functionally interchangeable and share an essential peptide motif. J. Virol. 1991, 65, 6001–6007. [Google Scholar] [CrossRef] [PubMed]
- Fischer, U.; Huber, J.; Boelens, W.C.; Mattaj, I.W.; Luhrmann, R. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell 1995, 82, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Meinkoth, J.L.; Tsien, R.Y.; Taylor, S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell 1995, 82, 463–473. [Google Scholar] [CrossRef]
- Meyer, B.E.; Malim, M.H. The HIV-1 Rev trans-activator shuttles between the nucleus and the cytoplasm. Genes Dev. 1994, 8424, 1538–1547. [Google Scholar] [CrossRef]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. Crm1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef]
- Neville, M.; Stutz, F.; Lee, L.; Davis, L.I.; Rosbash, M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr. Biol. 1997, 7, 767–775. [Google Scholar] [CrossRef]
- Ossareh-Nazari, B.; Bachelerie, F.; Dargemont, C. Evidence for a role of Crm1 in signal-mediated nuclear protein export. Science 1997, 278, 141–144. [Google Scholar] [CrossRef]
- Fukuda, M.; Asano, S.; Nakamura, T.; Adachi, M.; Yoshida, M.; Yanagida, M.; Nishida, E. Crm1 is responsible for intracellular transport mediated by the nuclear export signal. Nature 1997, 390, 308–311. [Google Scholar] [CrossRef]
- Hammarskjöld, M.-L. Regulation of retroviral rna export. Semin. Cell Dev. Biol. 1997, 8, 83–90. [Google Scholar] [CrossRef]
- Purcell, D.F.; Martin, M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993, 67, 6365–6378. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Bor, Y.C.; Misawa, Y.; Xue, Y.; Rekosh, D.; Hammarskjold, M.L. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature 2006, 443, 234–237. [Google Scholar] [CrossRef] [PubMed]
- LeBlanc, J.J.U.S.; Abraham, B.; Clatterbuck, S.; Beemon, K.L. Tap and Dbp5, but not Gag, are involved in DR-mediated nuclear export of unspliced rous sarcoma virus RNA. Virology 2007, 363, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Bray, M.; Prasad, S.; Dubay, J.W.; Hunter, E.; Jeang, K.T.; Rekosh, D.; Hammarskjold, M.L. A small element from the Mason-Pfizer monkey virus genome makes human immunodeficiency virus type 1 expression and replication Rev-independent. Proc. Nat. Acad. Sci. U. S. A. 1994, 91, 1256–1260. [Google Scholar] [CrossRef] [PubMed]
- Ernst, R.; Bray, M.; Rekosh, D.; Hammarskjöld, M.-L. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol. Cell. Biol. 1997, 17, 135–144. [Google Scholar] [CrossRef]
- Hidaka, M.; Inoue, J.; Yoshida, M.; Seiki, M. Post-transcriptional regulator (Rex) of HTLV-1 initiates expression of viral structural proteins but suppresses expression of regulatory proteins. EMBO J. 1988, 7, 519–523. [Google Scholar] [CrossRef]
- Seiki, M.; Inoue, J.; Hidaka, M.; Yoshida, M. Two cis-acting elements responsible for posttranscriptional trans-regulation of gene expression of human T-cell leukemia virus type 1. Proc. Nat. Acad. Sci. U. S. A. 1988, 85, 7124–7128. [Google Scholar] [CrossRef]
- Mertz, J.A.; Simper, M.S.; Lozano, M.M.; Payne, S.M.; Dudley, J.P. Mouse mammary tumor virus encodes a self-regulatory RNA export protein and is a complex retrovirus. J. Virol. 2005, 79, 14737–14747. [Google Scholar] [CrossRef]
- Martarano, L.; Stephens, R.; Rice, N.; Derse, D. Equine infectious anemia virus trans-regulatory protein Rev controls viral mRNA stability, accumulation, and alternative splicing. J. Virol. 1994, 68, 3102–3111. [Google Scholar] [CrossRef]
- Fridell, R.A.; Partin, K.M.; Carpenter, S.; Cullen, B.R. Identification of the activation domain of equine infectious anemia virus Rev. J. Virol. 1993, 67, 7317–7323. [Google Scholar] [CrossRef]
- Caporale, M.; Arnaud, F.; Mura, M.; Golder, M.; Murgia, C.; Palmarini, M. The signal peptide of a simple retrovirus envelope functions as a posttranscriptional regulator of viral gene expression. J. Virol. 2009, 83, 4591–4604. [Google Scholar] [CrossRef] [PubMed]
- Hofacre, A.; Nitta, T.; Fan, H. Jaagsiekte sheep retrovirus encodes a regulatory factor, Rej, required for synthesis of Gag protein. J. Virol. 2009, 83, 12483–12498. [Google Scholar] [CrossRef] [PubMed]
- Nitta, T.; Hofacre, A.; Hull, S.; Fan, H. Identification and mutational analysis of a Rej response element in Jaagsiekte sheep retrovirus RNA. J. Virol. 2009, 83, 12499–12511. [Google Scholar] [CrossRef] [PubMed]
- Bodem, J.; Schied, T.; Gabriel, R.; Rammling, M.; Rethwilm, A. Foamy virus nuclear RNA export is distinct from that of other retroviruses. J. Virol. 2011, 85, 2333–2341. [Google Scholar] [CrossRef] [PubMed]
- Cook, K.S.; Fisk, G.J.; Hauber, J.; Usman, N.; Daly, T.J.; Rusche, J.R. Characterization of HIV-1 Rev protein: Binding stoichiometry and minimal RNA substrate. Nucleic Acids Res. 1991, 19, 1577–1583. [Google Scholar] [CrossRef]
- Kjems, J.; Brown, M.; Chang, D.D.; Sharp, P.A. Structural analysis of the interaction between the human immunodeficiency virus rev protein and the Rev response element. Proc. Nat. Acad. Sci. U. S. A. 1991, 88, 683–687. [Google Scholar] [CrossRef]
- Madore, S.J.; Tiley, L.S.; Malim, M.H.; Cullen, B.R. Sequence requirements for Rev multimerization in vivo. Virology 1994, 202, 186–194. [Google Scholar] [CrossRef]
- Zapp, M.L.; Hope, T.J.; Parslow, T.G.; Green, M.R. Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: A dual function for an arginine-rich binding motif. Proc. Nat. Acad. Sci. U. S. A. 1991, 88, 7734–7748. [Google Scholar] [CrossRef]
- Olsen, H.S.; Cochrane, A.W.; Dillon, P.J.; Nalin, C.M.; Rosen, C.A. Interaction of the human immunodeficiency virus type 1 Rev protein with a structured region in Env mRNA is dependent on multimer formation mediated through a basic stretch of amino acids. Genes Dev. 1990, 4, 1357–1364. [Google Scholar] [CrossRef]
- Pond, S.J.; Ridgeway, W.K.; Robertson, R.; Wang, J.; Millar, D.P. HIV-1 Rev protein assembles on viral RNA one molecule at a time. Proc. Nat. Acad. Sci. U. S. A. 2009, 106, 1404–1408. [Google Scholar] [CrossRef]
- Charpentier, B.; Stutz, F.; Rosbash, M. A dynamic in vivo view of the HIV-1 Rev-RRE interaction. J. Mol. Biol. 1997, 266, 950–962. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P.; Zapp, M.L.; Green, M.R.; Szostak, J.W. HIV-1 Rev regulation involves recognition of non-watson-crick base pairs in viral RNA. Cell 1991, 67, 529–536. [Google Scholar] [CrossRef]
- Battiste, J.L.; Tan, R.; Frankel, A.D.; Williamson, J.R. Binding of an HIV Rev peptide to Rev responsive element RNA induces formation of purine-purine base pairs. Biochemistry 1994, 33, 2741–2747. [Google Scholar] [CrossRef] [PubMed]
- Battiste, J.L.; Mao, H.; Rao, N.S.; Tan, R.; Muhandiram, D.R.; Kay, L.E.; Frankel, A.D.; Williamson, J.R. Alpha helix-RNA major groove recognition in an HIV-1 Rev peptide-RRE RNA complex. Science 1996, 273, 1547–1551. [Google Scholar] [CrossRef] [PubMed]
- Tan, R.; Frankel, A.D. Costabilization of peptide and RNA structure in an HIV Rev peptide-RRE complex. Biochemistry 1994, 33, 14579–14585. [Google Scholar] [CrossRef]
- Cole, J.L.; Gehman, J.D.; Shafer, J.A.; Kuo, L.C. Solution oligomerization of the Rev protein of HIV-1. Biochemistry 1993, 32, 11769–11775. [Google Scholar] [CrossRef]
- Wingfield, P.T.; Stahl, S.J.; Payton, M.A.; Venkatesan, S.; Misra, M.; Steven, A.C. HIV-1 Rev expressed in recombinant escherichia coli: Purification, polymerization, and conformational properties. Biochemistry 1991, 30, 7527–7534. [Google Scholar] [CrossRef]
- Heaphy, S.; Finch, J.T.; Gait, M.J.; Karn, J.; Singh, M. Human immunodeficiency virus type 1 regulator of virion expression, Rev, forms nucleoprotein filaments after binding to a purine-rich “bubble” located within the Rev-responsive region of viral mRNAs. Proc. Nat. Acad. Sci. U. S. A. 1991, 88, 7366–7370. [Google Scholar] [CrossRef]
- Jain, C.; Belasco, J.G. A structural model for the HIV-1 Rev-RRE complex deduced from altered-specificity Rev variants isolated by a rapid genetic strategy. Cell 1996, 87, 115–125. [Google Scholar] [CrossRef]
- Jain, C.; Belasco, J.G. Structural model for the cooperative assembly of HIV-1 Rev multimers on the RRE as deduced from analysis of assembly-defective mutants. Mol. Cell 2001, 7, 603–614. [Google Scholar] [CrossRef]
- Zhou, P.; Wagner, G. Overcoming the solubility limit with solubility enhancement tags: Successful applications in biomolecular nmr studies. J. Biomol. NMR 2009, 46, 23–31. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2011 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Hammarskjold, M.-L.; Rekosh, D. A Long-Awaited Structure Is Rev-ealed. Viruses 2011, 3, 484-492. https://doi.org/10.3390/v3050484
Hammarskjold M-L, Rekosh D. A Long-Awaited Structure Is Rev-ealed. Viruses. 2011; 3(5):484-492. https://doi.org/10.3390/v3050484
Chicago/Turabian StyleHammarskjold, Marie-Louise, and David Rekosh. 2011. "A Long-Awaited Structure Is Rev-ealed" Viruses 3, no. 5: 484-492. https://doi.org/10.3390/v3050484