The Coronavirus E Protein: Assembly and Beyond
Abstract
:1. Introduction
2. Background

3. CoV E and Virus Assembly
4. Post-Translational Modifications
5. CoV E and the Cell Stress Response
6. Protein-Protein Interactions
7. The Ion Channel Activity of CoV E
8. Role of CoV E in Virion Release
8.1. The Mammalian Secretory Pathway and Large Cargo
8.2. Impact of CoV Infection on the Secretory Pathway
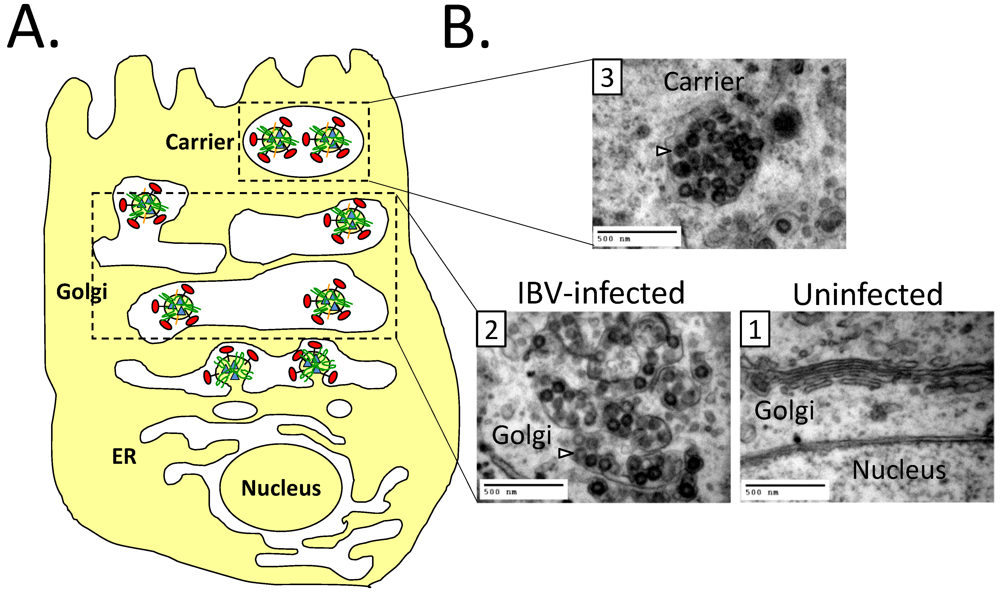
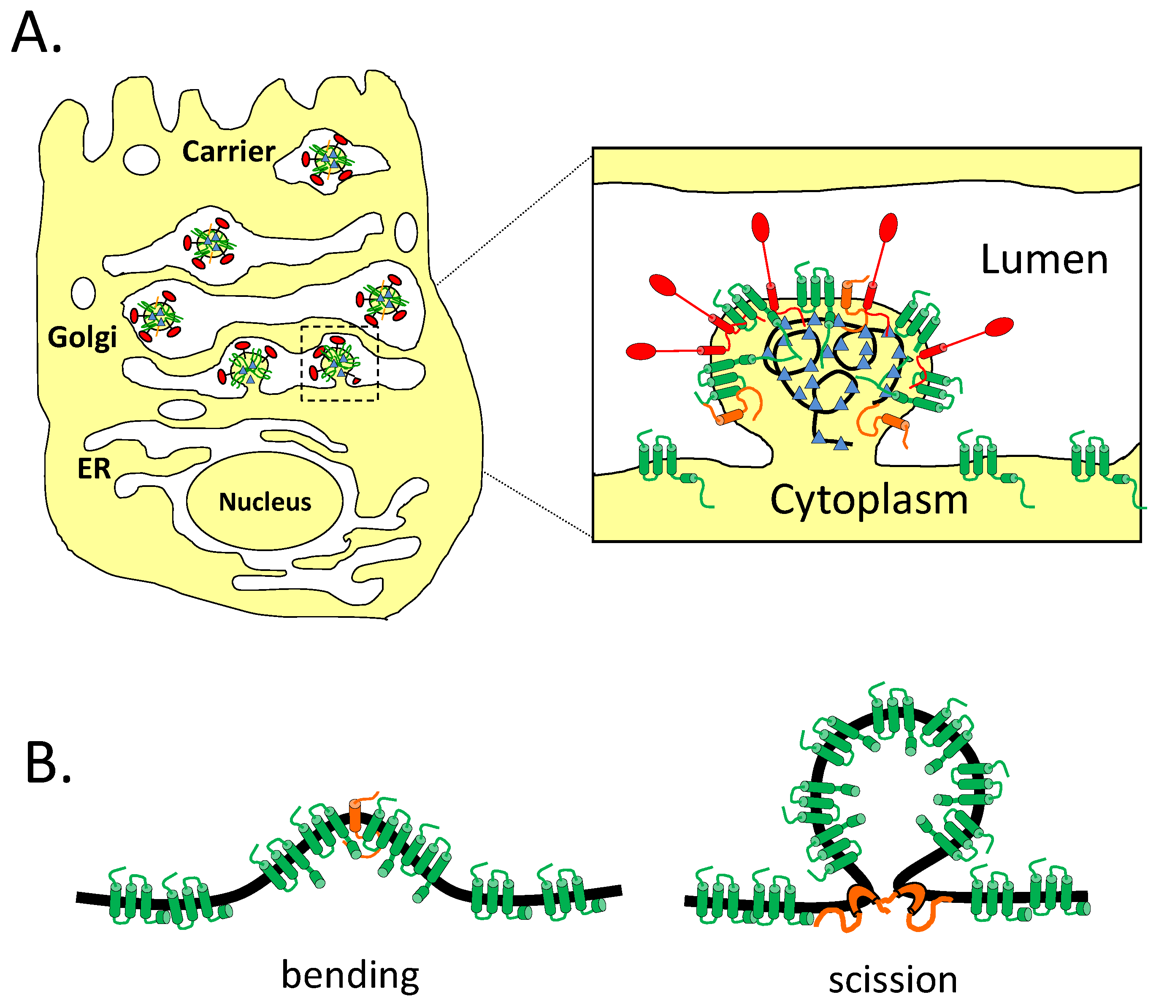
8.3. Effect of CoV E on the Secretory Pathway and Role in Release of Virions
9. Topology of CoV E
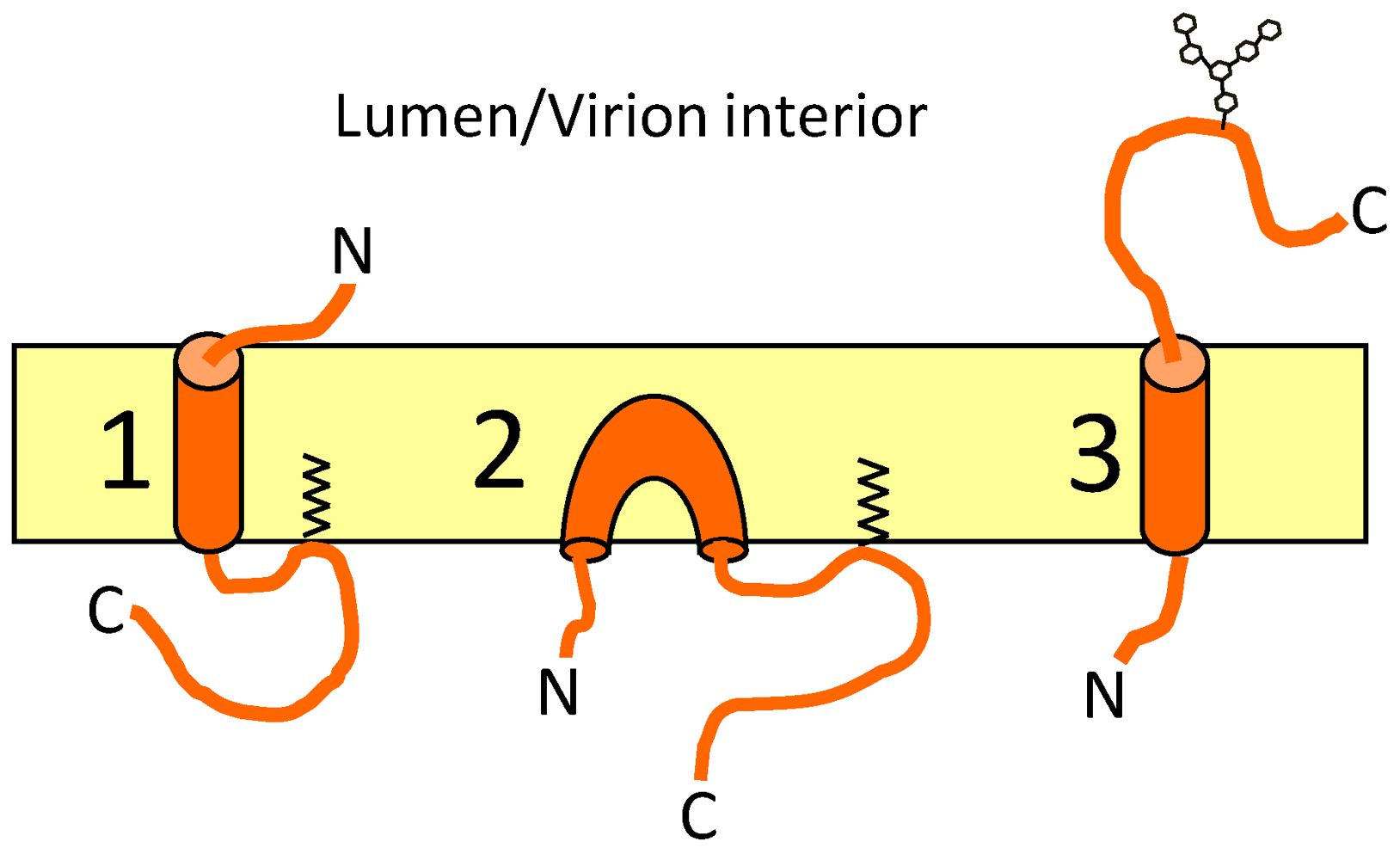
| Program | IBV E | MHV E | SARS E | TGEV E | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N | C | TMs | N | C | TMs | N | C | TMs | N | C | TMs | |
| TM Pred | lumen | lumen | 2 | lumen | cyto | 1 | lumen | cyto | 1 | lumen | cyto | 1 |
| HMMTop | lumen | lumen | 2 | cyto | cyto | 2 | lumen | cyto | 1 | cyto | cyto | 2 |
| TMHMM 2.0 | lumen | lumen | 2 | lumen | cyto | 1 | cyto | lumen | 1 | lumen | cyto | 1 |
| MEMSAT-SVM | lumen | lumen | 2 | lumen | lumen | 2 | lumen | lumen | 2 | cyto | lumen | 1 |
| MEMSAT3 | cyto | cyto | 2 | lumen | cyto | 1 | lumen | lumen | 2 | lumen | cyto | 1 |
10. Perspectives on CoV E
Acknowledgments
Conflict of Interest
References and Notes
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus. Res. 2006, 66, 193–292. [Google Scholar]
- Hogue, B.G.; Machamer, C.E. Coronavirus structural proteins and virus assembly. In Nidoviruses; Perlman, S., Gallagher, T., Snijder, E.J., Eds.; ASM Press: Washington, DC, USA, 2008; pp. 179–200. [Google Scholar]
- Boursnell, M.E.; Binns, M.M.; Brown, T.D. Sequencing of coronavirus IBV genomic RNA: Three open reading frames in the 5' 'unique' region of mRNA D. J. Gen. Virol. 1985, 66, 2253–2258. [Google Scholar]
- Budzilowicz, C.J.; Weiss, S.R. In vitro synthesis of two polypeptides from a nonstructural gene of coronavirus mouse hepatitis virus strain A59. Virology 1987, 157, 509–515. [Google Scholar] [CrossRef]
- Corse, E.; Machamer, C.E. Infectious bronchitis virus E protein is targeted to the Golgi complex and directs release of virus-like particles. J. Virol. 2000, 74, 4319–4326. [Google Scholar]
- Raamsman, M.J.; Locker, J.K.; de Hooge, A.; de Vries, A.A.; Griffiths, G.; Vennema, H.; Rottier, P.J. Characterization of the coronavirus mouse hepatitis virus strain A59 small membrane protein E. J. Virol. 2000, 74, 2333–2342. [Google Scholar]
- Liao, Y.; Yuan, Q.; Torres, J.; Tam, J.P.; Liu, D.X. Biochemical and functional characterization of the membrane association and membrane permeabilizing activity of the severe acute respiratory syndrome coronavirus envelope protein. Virology 2006, 349, 264–275. [Google Scholar]
- Cohen, J.R.; Lin, L.D.; Machamer, C.E. Identification of a Golgi complex-targeting signal in the cytoplasmic tail of the severe acute respiratory syndrome coronavirus envelope protein. J. Virol. 2011, 85, 5794–5803. [Google Scholar]
- Nieto-Torres, J.L.; Dediego, M.L.; Alvarez, E.; Jimenez-Guardeno, J.M.; Regla-Nava, J.A.; Llorente, M.; Kremer, L.; Shuo, S.; Enjuanes, L. Subcellular location and topology of severe acute respiratory syndrome coronavirus envelope protein. Virology 2011, 415, 69–82. [Google Scholar]
- Yuan, Q.; Liao, Y.; Torres, J.; Tam, J.P.; Liu, D.X. Biochemical evidence for the presence of mixed membrane topologies of the severe acute respiratory syndrome coronavirus envelope protein expressed in mammalian cells. FEBS Lett. 2006, 580, 3192–3200. [Google Scholar]
- Maeda, J.; Repass, J.F.; Maeda, A.; Makino, S. Membrane topology of coronavirus E protein. Virology 2001, 281, 163–169. [Google Scholar]
- Godet, M.; L'Haridon, R.; Vautherot, J.F.; Laude, H. TGEV corona virus ORF4 encodes a membrane protein that is incorporated into virions. Virology 1992, 188, 666–675. [Google Scholar]
- Corse, E.; Machamer, C.E. The cytoplasmic tail of infectious bronchitis virus E protein directs Golgi targeting. J. Virol. 2002, 76, 1273–1284. [Google Scholar]
- Boscarino, J.A.; Logan, H.L.; Lacny, J.J.; Gallagher, T.M. Envelope protein palmitoylations are crucial for murine coronavirus assembly. J. Virol. 2008, 82, 2989–2999. [Google Scholar]
- Lopez, L.A.; Riffle, A.J.; Pike, S.L.; Gardner, D.; Hogue, B.G. Importance of conserved cysteine residues in the coronavirus envelope protein. J. Virol. 2008, 82, 3000–3010. [Google Scholar]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar]
- Tooze, J.; Tooze, S.; Warren, G. Replication of coronavirus MHV-A59 in sac- cells: Determination of the first site of budding of progeny virions. Eur. J. Cell Biol. 1984, 33, 281–293. [Google Scholar]
- Klumperman, J.; Locker, J.K.; Meijer, A.; Horzinek, M.C.; Geuze, H.J.; Rottier, P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994, 68, 6523–6534. [Google Scholar]
- Vennema, H.; Godeke, G.J.; Rossen, J.W.; Voorhout, W.F.; Horzinek, M.C.; Opstelten, D.J.; Rottier, P.J. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996, 15, 2020–2028. [Google Scholar]
- Baudoux, P.; Carrat, C.; Besnardeau, L.; Charley, B.; Laude, H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J. Virol. 1998, 72, 8636–8643. [Google Scholar]
- Huang, Y.; Yang, Z.Y.; Kong, W.P.; Nabel, G.J. Generation of synthetic severe acute respiratory syndrome coronavirus pseudoparticles: Implications for assembly and vaccine production. J. Virol. 2004, 78, 12557–12565. [Google Scholar]
- Hsieh, P.K.; Chang, S.C.; Huang, C.C.; Lee, T.T.; Hsiao, C.W.; Kou, Y.H.; Chen, I.Y.; Chang, C.K.; Huang, T.H.; Chang, M.F. Assembly of severe acute respiratory syndrome coronavirus RNA packaging signal into virus-like particles is nucleocapsid dependent. J. Virol. 2005, 79, 13848–13855. [Google Scholar]
- Tseng, Y.T.; Wang, S.M.; Huang, K.J.; Lee, A.I.; Chiang, C.C.; Wang, C.T. Self-assembly of severe acute respiratory syndrome coronavirus membrane protein. J. Biol. Chem. 2010, 285, 12862–12872. [Google Scholar]
- Fang, Y.; Wu, N.; Gan, X.; Yan, W.; Morrell, J.C.; Gould, S.J. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007, 5, e158. [Google Scholar]
- Siu, Y.L.; Teoh, K.T.; Lo, J.; Chan, C.M.; Kien, F.; Escriou, N.; Tsao, S.W.; Nicholls, J.M.; Altmeyer, R.; Peiris, J.S.; et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J. Virol. 2008, 82, 11318–11330. [Google Scholar]
- Ruch, T.R.; Machamer, C.E. Johns Hopkins School of Medicine: Baltimore, MD, USA, 2010; Unpublished Work.
- Lim, K.P.; Liu, D.X. The missing link in coronavirus assembly. Retention of the avian coronavirus infectious bronchitis virus envelope protein in the pre-Golgi compartments and physical interaction between the envelope and membrane proteins. J. Biol. Chem. 2001, 276, 17515–17523. [Google Scholar]
- Corse, E.; Machamer, C.E. The cytoplasmic tails of infectious bronchitis virus E and M proteins mediate their interaction. Virology 2003, 312, 25–34. [Google Scholar]
- Fischer, F.; Stegen, C.F.; Masters, P.S.; Samsonoff, W.A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J. Virol. 1998, 72, 7885–7894. [Google Scholar]
- Ruch, T.R.; Machamer, C.E. The hydrophobic domain of infectious bronchitis virus E protein alters the host secretory pathway and is important for release of infectious virus. J. Virol. 2011, 85, 675–685. [Google Scholar]
- Arbely, E.; Khattari, Z.; Brotons, G.; Akkawi, M.; Salditt, T.; Arkin, I.T. A highly unusual palindromic transmembrane helical hairpin formed by SARS coronavirus E protein. J. Mol. Biol. 2004, 341, 769–779. [Google Scholar]
- Torres, J.; Parthasarathy, K.; Lin, X.; Saravanan, R.; Kukol, A.; Liu, D.X. Model of a putative pore: The pentameric alpha-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 2006, 91, 938–947. [Google Scholar]
- Kuo, L.; Masters, P.S. The small envelope protein E is not essential for murine coronavirus replication. J. Virol. 2003, 77, 4597–4608. [Google Scholar]
- DeDiego, M.L.; Alvarez, E.; Almazan, F.; Rejas, M.T.; Lamirande, E.; Roberts, A.; Shieh, W.J.; Zaki, S.R.; Subbarao, K.; Enjuanes, L. A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J. Virol. 2007, 81, 1701–1713. [Google Scholar]
- Ortego, J.; Ceriani, J.E.; Patino, C.; Plana, J.; Enjuanes, L. Absence of E protein arrests transmissible gastroenteritis coronavirus maturation in the secretory pathway. Virology 2007, 25, 296–308. [Google Scholar]
- Kuo, L.; Hurst, K.R.; Masters, P.S. Exceptional flexibility in the sequence requirements for coronavirus small envelope protein function. J. Virol. 2007, 81, 2249–2262. [Google Scholar]
- Kuo, L.; Masters, P.S. Evolved variants of the membrane protein can partially replace the envelope protein in murine coronavirus assembly. J. Virol. 2010, 84, 12872–12885. [Google Scholar]
- Curtis, K.M.; Yount, B.; Baric, R.S. Heterologous gene expression from transmissible gastroenteritis virus replicon particles. J. Virol. 2002, 76, 1422–1434. [Google Scholar]
- Ortego, J.; Escors, D.; Laude, H.; Enjuanes, L. Generation of a replication-competent, propagation-deficient virus vector based on the transmissible gastroenteritis coronavirus genome. J. Virol. 2002, 76, 11518–11529. [Google Scholar]
- Alvarez, E.; DeDiego, M.L.; Nieto-Torres, J.L.; Jimenez-Guardeno, J.M.; Marcos-Villar, L.; Enjuanes, L. The envelope protein of severe acute respiratory syndrome coronavirus interacts with the non-structural protein 3 and is ubiquitinated. Virology 2010, 402, 281–291. [Google Scholar]
- Keng, C.T.; Akerstrom, S.; Leung, C.S.; Poon, L.L.; Peiris, J.S.; Mirazimi, A.; Tan, Y.J. SARS coronavirus 8b reduces viral replication by down-regulating E via an ubiquitin-independent proteasome pathway. Microb. Infect. 2011, 13, 179–188. [Google Scholar]
- An, S.; Chen, C.J.; Yu, X.; Leibowitz, J.L.; Makino, S. Induction of apoptosis in murine coronavirus-infected cultured cells and demonstration of E protein as an apoptosis inducer. J. Virol. 1999, 73, 7853–7859. [Google Scholar]
- Yang, Y.; Xiong, Z.; Zhang, S.; Yan, Y.; Nguyen, J.; Ng, B.; Lu, H.; Brendese, J.; Yang, F.; Wang, H.; et al. Bcl-xL inhibits T-cell apoptosis induced by expression of SARS coronavirus E protein in the absence of growth factors. Biochem. J. 2005, 392, 135–143. [Google Scholar] [CrossRef]
- Dediego, M.L.; Nieto-Torres, J.L.; Jimenez-Guardeno, J.M.; Regla-Nava, J.A.; Alvarez, E.; Oliveros, J.C.; Zhao, J.; Fett, C.; Perlman, S.; Enjuanes, L. Severe acute respiratory syndrome coronavirus envelope protein regulates cell stress response and apoptosis. PLoS Pathog. 2011, 7, e1002315. [Google Scholar]
- Teoh, K.T.; Siu, Y.L.; Chan, W.L.; Schluter, M.A.; Liu, C.J.; Peiris, J.S.; Bruzzone, R.; Margolis, B.; Nal, B. The SARS coronavirus E protein interacts with PALS1 and alters tight junction formation and epithelial morphogenesis. Mol. Biol. Cell 2010, 21, 3838–3852. [Google Scholar]
- Griffin, S.D.; Beales, L.P.; Clarke, D.S.; Worsfold, O.; Evans, S.D.; Jaeger, J.; Harris, M.P.; Rowlands, D.J. The p7 protein of hepatitis C virus forms an ion channel that is blocked by the antiviral drug, Amantadine. FEBS Lett. 2003, 535, 34–38. [Google Scholar]
- Pavlovic, D.; Neville, D.C.; Argaud, O.; Blumberg, B.; Dwek, R.A.; Fischer, W.B.; Zitzmann, N. The hepatitis C virus p7 protein forms an ion channel that is inhibited by long-alkyl-chain iminosugar derivatives. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 6104–6108. [Google Scholar]
- Pinto, L.H.; Holsinger, L.J.; Lamb, R.A. Influenza virus M2 protein has ion channel activity. Cell 1992, 69, 517–528. [Google Scholar]
- Aldabe, R.; Barco, A.; Carrasco, L. Membrane permeabilization by poliovirus proteins 2B and 2BC. J. Biol. Chem. 1996, 271, 23134–23137. [Google Scholar]
- Wang, K.; Xie, S.Q.; Sun, B. Viral proteins function as ion channels. Biochim. Biophys. Acta Biomembr. 2011, 1808, 510–515. [Google Scholar]
- Wharton, S.A.; Belshe, R.B.; Skehel, J.J.; Hay, A.J. Role of virion M2 protein in influenza virus uncoating: specific reduction in the rate of membrane fusion between virus and liposomes by amantadine. J. Gen. Virol. 1994, 75, 945–948. [Google Scholar]
- Sakaguchi, T.; Leser, G.P.; Lamb, R.A. The ion channel activity of the influenza virus M2 protein affects transport through the Golgi apparatus. J. Cell Biol. 1996, 133, 733–747. [Google Scholar]
- Henkel, J.R.; Weisz, O.A. Influenza virus M2 protein slows traffic along the secretory pathway. pH perturbation of acidified compartments affects early Golgi transport steps. J. Biol. Chem. 1998, 273, 6518–6524. [Google Scholar] [CrossRef]
- Ciampor, F.; Bayley, P.M.; Nermut, M.V.; Hirst, E.M.; Sugrue, R.J.; Hay, A.J. Evidence that the amantadine-induced, M2-mediated conversion of influenza A virus hemagglutinin to the low pH conformation occurs in an acidic trans Golgi compartment. Virology 1992, 188, 14–24. [Google Scholar]
- Carrere-Kremer, S.; Montpellier-Pala, C.; Cocquerel, L.; Wychowski, C.; Penin, F.; Dubuisson, J. Subcellular localization and topology of the p7 polypeptide of hepatitis C virus. J. Virol. 2002, 76, 3720–3730. [Google Scholar]
- Wozniak, A.L.; Griffin, S.; Rowlands, D.; Harris, M.; Yi, M.; Lemon, S.M.; Weinman, S.A. Intracellular proton conductance of the hepatitis C virus p7 protein and its contribution to infectious virus production. PLoS Pathog. 2010, 6, e1001087. [Google Scholar]
- Steinmann, E.; Penin, F.; Kallis, S.; Patel, A.H.; Bartenschlager, R.; Pietschmann, T. Hepatitis C virus p7 protein is crucial for assembly and release of infectious virions. PLoS Pathog. 2007, 3, e103. [Google Scholar]
- Brohm, C.; Steinmann, E.; Friesland, M.; Lorenz, I.C.; Patel, A.; Penin, F.; Bartenschlager, R.; Pietschmann, T. Characterization of determinants important for hepatitis C virus p7 function in morphogenesis by using trans-complementation. J. Virol. 2009, 83, 11682–11693. [Google Scholar]
- Wilson, L.; McKinlay, C.; Gage, P.; Ewart, G. SARS coronavirus E protein forms cation-selective ion channels. Virology 2004, 330, 322–331. [Google Scholar]
- Wilson, L.; Gage, P.; Ewart, G. Hexamethylene amiloride blocks E protein ion channels and inhibits coronavirus replication. Virology 2006, 353, 294–306. [Google Scholar]
- Torres, J.; Wang, J.; Parthasarathy, K.; Liu, D.X. The transmembrane oligomers of coronavirus protein E. Biophys. J. 2005, 88, 1283–1290. [Google Scholar]
- Torres, J.; Maheswari, U.; Parthasarathy, K.; Ng, L.; Liu, D.X.; Gong, X. Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein Sci. 2007, 16, 2065–2071. [Google Scholar]
- Parthasarathy, K.; Ng, L.; Lin, X.; Liu, D.X.; Pervushin, K.; Gong, X.; Torres, J. Structural flexibility of the pentameric SARS coronavirus envelope protein ion channel. Biophys. J. 2008, 95, L39–41. [Google Scholar]
- Pervushin, K.; Tan, E.; Parthasarathy, K.; Lin, X.; Jiang, F.L.; Yu, D.; Vararattanavech, A.; Soong, T.W.; Liu, D.X.; Torres, J. Structure and inhibition of the SARS coronavirus envelope protein ion channel. PLoS Pathog. 2009, 5, e1000511. [Google Scholar]
- Ruch, T.R.; Machamer, C.E. A Single polar residue and distinct membrane topologies impact the function of the infectious bronchitis coronavirus E protein. PLoS Pathog. 2012. submitted for publication. [Google Scholar]
- Ye, Y.; Hogue, B.G. Role of the coronavirus E viroporin protein transmembrane domain in virus assembly. J. Virol. 2007, 81, 3597–3607. [Google Scholar]
- Appenzeller-Herzog, C.; Hauri, H.P. The ER-Golgi intermediate compartment (ERGIC): In search of its identity and function. J. Cell Sci. 2006, 119, 2173–2183. [Google Scholar]
- Klumperman, J. Architecture of the mammalian Golgi. Cold Spring Harb. Perspect. Biol. 2011, 3. [Google Scholar]
- Marsh, B.J.; Howell, K.E. The mammalian Golgi—Complex debates. Nat. Rev. Mol. Cell Biol. 2002, 3, 789–795. [Google Scholar]
- Morre, D.J.; Ovtracht, L. Dynamics of the Golgi apparatus: Membrane differentiation and membrane flow. Int. Rev. Cytol. Suppl. 1977, 5, 61–188. [Google Scholar]
- Balch, W.E.; Dunphy, W.G.; Braell, W.A.; Rothman, J.E. Reconstitution of the transport of protein between successive compartments of the Golgi measured by the coupled incorporation of N-acetylglucosamine. Cell 1984, 39, 405–416. [Google Scholar]
- Balch, W.E.; Glick, B.S.; Rothman, J.E. Sequential intermediates in the pathway of intercompartmental transport in a cell-free system. Cell 1984, 39, 525–536. [Google Scholar]
- Bonfanti, L.; Mironov, A.A., Jr.; Martinez-Menarguez, J.A.; Martella, O.; Fusella, A.; Baldassarre, M.; Buccione, R.; Geuze, H.J.; Mironov, A.A.; Luini, A. Procollagen traverses the Golgi stack without leaving the lumen of cisternae: evidence for cisternal maturation. Cell 1998, 95, 993–1003. [Google Scholar]
- Losev, E.; Reinke, C.A.; Jellen, J.; Strongin, D.E.; Bevis, B.J.; Glick, B.S. Golgi maturation visualized in living yeast. Nature 2006, 441, 1002–1006. [Google Scholar]
- Matsuura-Tokita, K.; Takeuchi, M.; Ichihara, A.; Mikuriya, K.; Nakano, A. Live imaging of yeast Golgi cisternal maturation. Nature 2006, 441, 1007–1010. [Google Scholar]
- Volchuk, A.; Amherdt, M.; Ravazzola, M.; Brugger, B.; Rivera, V.M.; Clackson, T.; Perrelet, A.; Sollner, T.H.; Rothman, J.E.; Orci, L. Megavesicles implicated in the rapid transport of intracisternal aggregates across the Golgi stack. Cell 2000, 102, 335–348. [Google Scholar]
- Mironov, A.A.; Beznoussenko, G.V.; Nicoziani, P.; Martella, O.; Trucco, A.; Kweon, H.S.; Di Giandomenico, D.; Polishchuk, R.S.; Fusella, A.; Lupetti, P.; et al. Small cargo proteins and large aggregates can traverse the Golgi by a common mechanism without leaving the lumen of cisternae. J. Cell Biol. 2001, 155, 1225–1238. [Google Scholar]
- Trelstad, R.L.; Hayashi, K. Tendon collagen fibrillogenesis: Intracellular subassemblies and cell surface changes associated with fibril growth. Dev. Biol. 1979, 71, 228–242. [Google Scholar]
- Leblond, C.P. Synthesis and secretion of collagen by cells of connective tissue, bone, and dentin. Anat. Rec. 1989, 224, 123–138. [Google Scholar] [CrossRef]
- Ueno, T.; Kaneko, K.; Katano, H.; Sato, Y.; Mazitschek, R.; Tanaka, K.; Hattori, S.; Irie, S.; Sata, T.; Ogawa-Goto, K. Expansion of the trans-Golgi network following activated collagen secretion is supported by a coiled-coil microtubule-bundling protein, p180, on the ER. Exp. Cell Res. 2010, 316, 329–340. [Google Scholar]
- Hussain, M.M. A proposed model for the assembly of chylomicrons. Atherosclerosis 2000, 148, 1–15. [Google Scholar]
- Mansbach, C.M.; Siddiqi, S.A. The biogenesis of chylomicrons. Annu. Rev. Physiol. 2010, 72, 315–333. [Google Scholar]
- Sabesin, S.M.; Frase, S. Electron microscopic studies of the assembly, intracellular transport, and secretion of chylomicrons by rat intestine. J. Lipid Res. 1977, 18, 496–511. [Google Scholar]
- Lavi, E.; Wang, Q.; Weiss, S.R.; Gonatas, N.K. Syncytia formation induced by coronavirus infection is associated with fragmentation and rearrangement of the Golgi apparatus. Virology 1996, 221, 325–334. [Google Scholar]
- Ulasli, M.; Verheije, M.H.; de Haan, C.A.; Reggiori, F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010, 12, 844–861. [Google Scholar]
- Mackenzie, J.M.; Jones, M.K.; Westaway, E.G. Markers for trans-Golgi membranes and the intermediate compartment localize to induced membranes with distinct replication functions in flavivirus-infected cells. J. Virol. 1999, 73, 9555–9567. [Google Scholar]
- Gillespie, L.K.; Hoenen, A.; Morgan, G.; Mackenzie, J.M. The endoplasmic reticulum provides the membrane platform for biogenesis of the flavivirus replication complex. J. Virol. 2010, 84, 10438–10447. [Google Scholar]
- Mackenzie, J.M.; Khromykh, A.A.; Parton, R.G. Cholesterol manipulation by West Nile virus perturbs the cellular immune response. Cell Host Microbe 2007, 2, 229–239. [Google Scholar]
- Griffiths, G.; Rottier, P. Cell biology of viruses that assemble along the biosynthetic pathway. Semin. Cell Biol. 1992, 3, 367–381. [Google Scholar]
- Murphy, F.A.; Harrison, A.K.; Whitfield, S.G. Bunyaviridae: Morphologic and morphogenetic similarities of Bunyamwera serologic supergroup viruses and several other arthropod-borne viruses. Intervirology 1973, 1, 297–316. [Google Scholar]
- Salanueva, I.J.; Novoa, R.R.; Cabezas, P.; Lopez-Iglesias, C.; Carrascosa, J.L.; Elliott, R.M.; Risco, C. Polymorphism and structural maturation of bunyamwera virus in Golgi and post-Golgi compartments. J. Virol. 2003, 77, 1368–1381. [Google Scholar]
- Hoffman, K.; Stoffel, W. TMBASE-A database of membrane spanning protein segments. Biol. Chem. 1993, 166. [Google Scholar]
- Tusnady, G.E.; Simon, I. Principles governing amino acid composition of integral membrane proteins: Application to topology prediction. J. Mol. Biol. 1998, 283, 489–506. [Google Scholar]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar]
- Jones, D.T. Improving the accuracy of transmembrane protein topology prediction using evolutionary information. Bioinformatics 2007, 23, 538–544. [Google Scholar]
- Nugent, T.; Jones, D.T. Transmembrane protein topology prediction using support vector machines. BMC Bioinformatics 2009, 10, 159. [Google Scholar]
- Parton, R.G.; Simons, K. The multiple faces of caveolae. Nat. Rev. Mol. Cell Biol. 2007, 8, 185–194. [Google Scholar]
- Monier, S.; Parton, R.G.; Vogel, F.; Behlke, J.; Henske, A.; Kurzchalia, T.V. VIP21-caveolin, a membrane protein constituent of the caveolar coat, oligomerizes in vivo and in vitro. Mol. Biol. Cell 1995, 6, 911–927. [Google Scholar]
- Borgese, N.; Colombo, S.; Pedrazzini, E. The tale of tail-anchored proteins: Coming from the cytosol and looking for a membrane. J. Cell Biol. 2003, 161, 1013–1019. [Google Scholar]
- Shao, S.; Hegde, R.S. A calmodulin-dependent translocation pathway for small secretory proteins. Cell 2011, 147, 1576–1588. [Google Scholar]
© 2012 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ruch, T.R.; Machamer, C.E. The Coronavirus E Protein: Assembly and Beyond. Viruses 2012, 4, 363-382. https://doi.org/10.3390/v4030363
Ruch TR, Machamer CE. The Coronavirus E Protein: Assembly and Beyond. Viruses. 2012; 4(3):363-382. https://doi.org/10.3390/v4030363
Chicago/Turabian StyleRuch, Travis R., and Carolyn E. Machamer. 2012. "The Coronavirus E Protein: Assembly and Beyond" Viruses 4, no. 3: 363-382. https://doi.org/10.3390/v4030363
APA StyleRuch, T. R., & Machamer, C. E. (2012). The Coronavirus E Protein: Assembly and Beyond. Viruses, 4(3), 363-382. https://doi.org/10.3390/v4030363





