Mutation Distribution in the NSP4 Protein in Rotaviruses Isolated from Mexican Children with Moderate to Severe Gastroenteritis
Abstract
:1. Introduction
2. Results and Discussion
2.1. Rotavirus Positive Samples and Gastroenteritis Severity
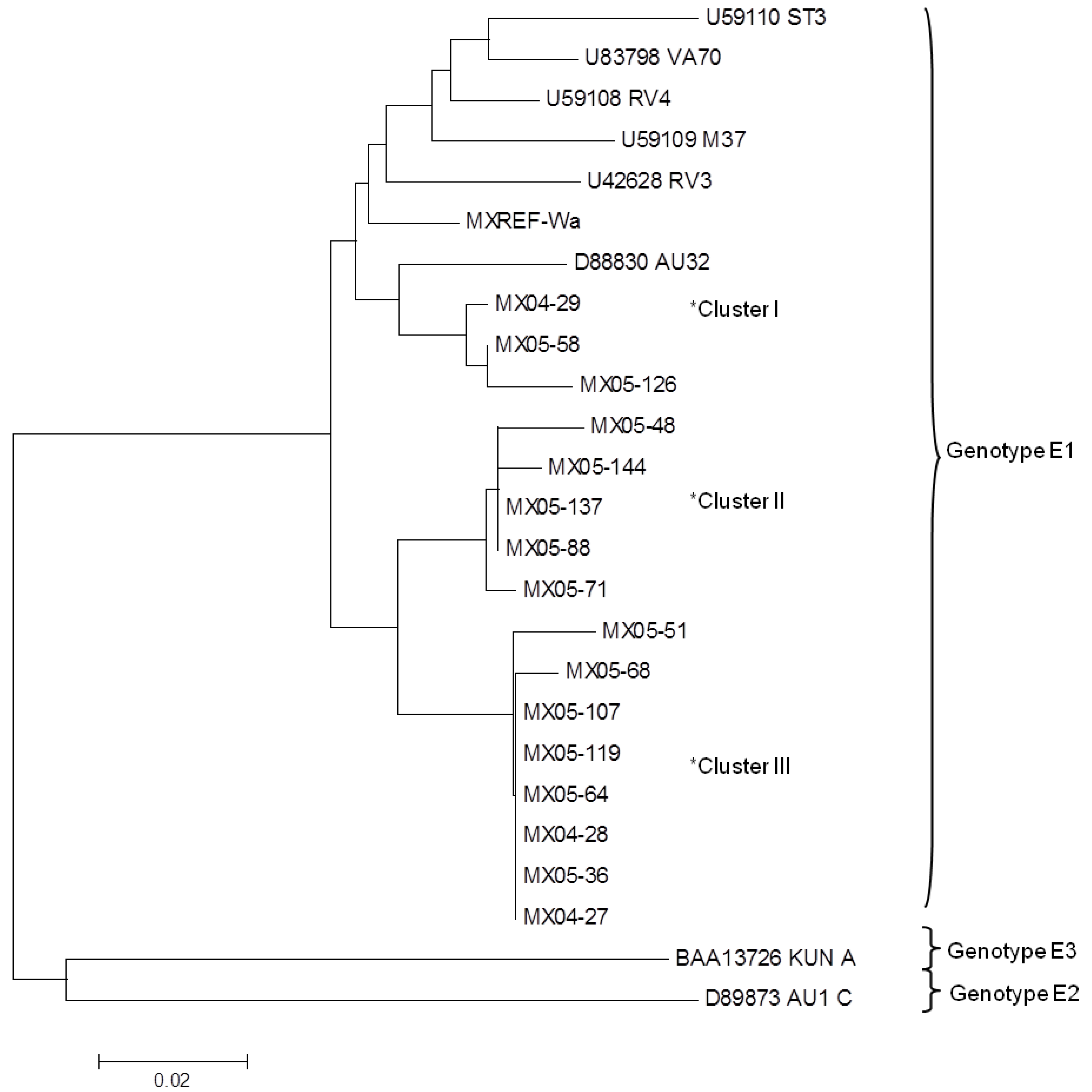
2.2. The NSP4 Genotype
2.3. The NSP4 Sequence Analysis
| Gastroenteritis severity (Ruuska score) | Gastroenteritis severity | Incidence | Breastfeeding | Sex | Age* (months) | diarrhea episodes* /24 h | Days with diarrhea* | Vomiting episodes* / 24hrs | Days of vomiting* | |
|---|---|---|---|---|---|---|---|---|---|---|
| M | F | |||||||||
| ≤ 10 | Mild | 13 (26%) | 61.5% | 6 | 7 | 8 | 6.1 | 2.4 | 2.6 | 1.0 |
| ≥ 11 | Moderate | 23 (46%) | 82.3% | 15 | 8 | 12 | 8.1 | 3.1 | 4.8 | 2.7 |
| ≥ 15 | Severe | 14 (28%) | 71.4% | 7 | 7 | 14 | 10.4 | 4.3 | 9.3 | 3.6 |
| Transmembrane site (22−44) | |||||||||||||||||||||||||
| GS1 | H1 (aa 7−21) | GS2 | |||||||||||||||||||||||
| Consensus seq | M | D | K | L | A | D | L | N | Y | T | L | S | V | I | T | L | M | N | D | T | L | H | S | I | I |
| Frequency % | 100 | 98.5 | 99.7 | 98 | 97.6 | 99.5 | 100 | 99.7 | 100 | 99.4 | 99.4 | 92.7 | 99.7 | 97.9 | 98.2 | 99 | 100 | 99.4 | 97.9 | 98.8 | 100 | 98.8 | 98.8 | 100 | 98.8 |
| aa position | 26 | 27 | 28 | 29 | 30 | 31 | 32 | 33 | 34 | 35 | 36 | 37 | 38 | 39 | 40 | 41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49 | 50 |
| H2 (29−47) | |||||||||||||||||||||||||
| Transmembrane domain (22−44) | |||||||||||||||||||||||||
| Consensus seq. | Q | D | P | G | M | A | Y | F | P | Y | I | A | S | V | L | T | V | L | F | T | L | H | K | A | S |
| Frequency % | 97.6 | 99.7 | 100 | 100 | 96.4 | 100 | 100 | 100 | 97.8 | 100 | 98.8 | 100 | 99.7 | 99.4 | 100 | 100 | 994 | 99.4 | 99.7 | 98.4 | 100 | 99.4 | 99.4 | 100 | 99.4 |
| aa position | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 |
| H3 ( aa 67-85) | |||||||||||||||||||||||||
| Consensus seq. | I | P | T | M | K | I | A | L | K | T | S | K | C | S | Y | K | V | I | K | Y | C | I | V | T | I |
| Frequency % | 99.4 | 99.4 | 99.1 | 99 | 99.7 | 99.4 | 100 | 99.4 | 96.6 | 99.4 | 100 | 98.8 | 99.1 | 100 | 99.7 | 100 | 99.4 | 97.9 | 99.1 | 98.5 | 98.8 | 88.9 | 99.1 | 99.4 | 95.5 |
| aa position | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90 | 91 | 92 | 93 | 94 | 95 | 96 | 97 | 98 | 99 | 100 |
| H3 (67−85) | Alpha coiled Domain (95−137) | ||||||||||||||||||||||||
| Consensus seq. | I | N | T | L | L | K | L | A | G | Y | K | E | Q | V | T | T | K | D | E | I | E | Q | Q | M | D |
| Frequency % | 87.4 | 99.7 | 98.5 | 100 | 99.1 | 98.8 | 98.2 | 98.8 | 100 | 99.4 | 98.5 | 100 | 99.1 | 96.7 | 98.8 | 98 | 99.1 | 98.5 | 98.8 | 99.7 | 100 | 98.2 | 99.7 | 99.7 | 99.1 |
| aa position | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 | 109 | 110 | 111 | 112 | 113 | 114 | 115 | 116 | 117 | 118 | 119 | 120 | 121 | 122 | 123 | 124 | 125 |
| Alpha coiled Domain (95−137) | |||||||||||||||||||||||||
| VP4 Binding site (112−148) | |||||||||||||||||||||||||
| Enterotoxin (114-135) | |||||||||||||||||||||||||
| Consensus seq. | R | I | V | K | E | M | R | R | Q | L | E | M | I | D | K | L | T | T | R | E | I | E | Q | V | E |
| Frequency % | 99.7 | 98.8 | 99.1 | 99 | 99.7 | 99.4 | 99.7 | 100 | 98.5 | 99.7 | 93.4 | 99.4 | 99.7 | 99.1 | 99.1 | 100 | 100 | 98.8 | 99.1 | 99.7 | 99.7 | 99.7 | 99.7 | 99.4 | 100 |
| aa position | 126 | 127 | 128 | 129 | 130 | 131 | 132 | 133 | 134 | 135 | 136 | 137 | 138 | 139 | 140 | 141 | 142 | 143 | 144 | 145 | 146 | 147 | 148 | 149 | 150 |
| Alpha coiled Domain (95−137) | |||||||||||||||||||||||||
| VP4 Binding site (112−148) | |||||||||||||||||||||||||
| Enterotoxin (114−135) | |||||||||||||||||||||||||
| Consensus seq. | L | L | K | R | I | H | D | N | L | I | T | R | P | V | D | V | I | D | M | S | K | E | F | N | Q |
| Frequency % | 99.4 | 100 | 99.7 | 99 | 99.7 | 92.8 | 99.1 | 78.6 | 100 | 90.7 | 93.4 | 87.1 | 78.4 | 88 | 89.8 | 58 | 76 | 99.4 | 97.9 | 64.3 | 99.1 | 99.1 | 97.3 | 99.1 | 100 |
| aa position | 151 | 152 | 153 | 154 | 155 | 156 | 157 | 158 | 159 | 160 | 161 | 162 | 163 | 164 | 165 | 166 | 167 | 168 | 169 | 170 | 171 | 172 | 173 | 174 | 175 |
| VP6 Binding site (167−175) | |||||||||||||||||||||||||
| Consensus seq. | K | N | I | K | T | L | D | E | W | E | S | G | K | N | P | Y | E | P | S | E | V | T | A | S | M |
| Frequency % | 98.8 | 99.7 | 94 | 93 | 100 | 99.7 | 98.2 | 99.1 | 100 | 95.8 | 77 | 99.7 | 97.6 | 99.1 | 100 | 100 | 99.1 | 100 | 56 | 99.1 | 99.4 | 99.7 | 100 | 98.8 | 100 |
| NSP4 Sequence | Severity scores | Cluster | Amino acid variability and distribution | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H3 * | TD | ACD * | E/VP4 * | VP4 * | VP6 * | |||||||||||||||
| 76 | 77 | 85 | 94 | 111 | 132 | 136 | 137 | 141 | 142 | 144 | 145 | 161 | 169 | 172 | 173 | 174 | 175 | |||
| MX04-29 | 16 | I | N | A | K | |||||||||||||||
| MX05-58 | 14 | I | D | A | K | |||||||||||||||
| MX05-126 | 16 | I | V | D | A | K | ||||||||||||||
| MX05-48 | 8 | II | V | H | T | T | N | I | ||||||||||||
| MX05-144 | 13 | II | V | T | T | N | I | |||||||||||||
| MX05-137 | 15 | II | V | T | T | N | I | |||||||||||||
| MX05-88 | 16 | II | V | T | T | N | I | |||||||||||||
| MX05-71 | 0 | II | V | T | T | N | M | |||||||||||||
| MX04-27 | 11 | III | S | V | V | T | I | |||||||||||||
| MX04-28 | 14 | III | S | V | V | T | I | |||||||||||||
| MX05-36 | 8 | III | S | V | V | T | I | |||||||||||||
| MX05-51 | 14 | III | N | S | V | V | T | I | I | |||||||||||
| MX05-64 | 12 | III | S | V | V | T | I | |||||||||||||
| MX05-68 | 8 | III | G | S | V | V | T | I | ||||||||||||
| MX05-107 | 14 | III | S | V | V | T | I | |||||||||||||
| MX05-119 | 15 | III | S | V | V | T | N | I | ||||||||||||
| Consensus amino acids | I | N | Y | E | E | D | T | R | V | I | M | S | S | S | T | A | S | M | ||
| Frequency % | 86 | 99.7 | 99.4 | 99.1 | 89.4 | 99.4 | 92.8 | 85.7 | 56.2 | 75.9 | 99.4 | 61 | 76 | 56 | 100 | 100 | 99 | 100 | ||
3. Experimental Section
3.1. Samples Recollection and Gastroenteritis Severity Score
3.2. Extraction of Rotavirus RNA.
3.3. NSP4 Genotype Identification and Sequence
4. Conclusions
Conflict of Interest
References and Notes
- Kapikian, A.; Hoshino, Y.; Chanock, R. Rotaviruses. In Fields virology, 4th; Knipe, D.M., Howley, P.M., Griffin, D.E., Lamb, R.A., Martin, M.A., Roizman, B., Strais, S.E., Eds.; Lippincott Williams and Wilkins: Philadelphia, Pennsylvania Pa, USA, 2001; pp. 1787–1833. [Google Scholar]
- Morris, A.P.; Estes, M.K. Microbes and microbial toxins: Paradigms for microbial mucosal interactions VIII: Pathological consequences of rotavirus infection an enterotoxin. Am. J. Physiol.-Gastr. L. 2001, 281, 303–310. [Google Scholar]
- Ramig, R.F. Pathogenesis of intestinal and systemic rotavirus infection. J. Virol. 2004, 78, 10213–10220. [Google Scholar] [CrossRef]
- Tate, J.E.; Burton, A.H.; Boschi-Pinto, C.; Steele, A.D.; Duque, J.; Parashar, U.D. 2008 estimate of worldwide rotavirus-associated mortality in children younger than 5 years before the introduction of universal rotavirus vaccination programmes: A systematic review and meta-analysis. Lancet Infect. Dis. 2012, 12, 131–141. [Google Scholar]
- Ball, J.M.; Tian, P.; Zeng, C.Q.; Morris, A.P.; Estes, M.K. Age-dependent diarrhea induced by a rotaviral nonstructural glycoprotein. Science 1996, 272, 101–104. [Google Scholar]
- Dong, Y.; Zeng, C.; Ball, J.; Estes, M.; Morris, A. The rotavirus enterotoxin NSP4 mobilizes intracellular calcium in human intestinal cells by stimulating phospholipase C-mediated inositol 1,4,5-trisphosphate production. Proc. Natl. Acad. Sci. USA 1997, 94, 3960–3965. [Google Scholar] [CrossRef]
- Brunet, J.P.; Cotte, J.; Linxe, C.; Quero, A.M; Géniteau-Legendre, M.; Servin, A. Rotavirus infection induces an increase in intracellular calcium concentration in human intestinal epithelial cells: Role in microvillar actin alteration. J. Virol. 2000, 74, 2323–2332. [Google Scholar]
- Zhang, M.; Zeng, C.Q.; Morris, A.P.; Estes, M.K. A functional NSP4 enterotoxin peptide secreted from rotavirus-infected cells. J. Virol. 2000, 74, 663–11670. [Google Scholar]
- Estes, M.K. Rotaviruses and their replication. In Fields virology, 4th; Knipe, D.M., Howley, P.M., Griffin, D.E., Lamb, R.A., Martin, M.A., Roizman, B., Strais, S.E., Eds.; Lippincott Williams and Wilkins: Philadelphia, Pennsylvania, USA, 2001; pp. 1747–1785. [Google Scholar]
- Au, K.; Chan, W.; Burns, J.W.; Estes, M.K. Receptor activity of rotavirus nonstructural glycoprotein NS28. J. Virol. 1989, 63, 4553–4562. [Google Scholar]
- Bergmann, C.C.; Maas, D.; Poruchynsky, M.S.; Atkinson, P.H.; Bellamy, A.R. Topology of the non-structural rotavirus receptor glycoprotein NS28 in the rough endoplasmic reticulum. EMBO J. 1989, 8, 1695–1703. [Google Scholar]
- Tian, P.; Ball, J.; Zeng, C.; Estes, M. Rotavirus Protein Expression is important for virus assembly and pathogenesis. Arch. Virol. 1996, 12, 69–77. [Google Scholar]
- Mirazimi, A.; Nilsson, M.; Svensson, L. The molecular chaperone calnexin interacts with the NSP4 enterotoxin of rotavirus in vivo and in vitro. J. Virol. 1998, 72, 8705–8709. [Google Scholar]
- Xu, A.; Bellamy, A.R.; Taylor, J.A. Immobilization of the early secretory pathway by a virus glycoprotein that binds to microtubules. EMBO J. 2000, 19, 6465–6474. [Google Scholar] [CrossRef]
- Storey, S.M.; Gibbons, T.F.; Williams, C.V.; Parr, R.D.; Schroeder, F.; Ball, J.M. Full-length, glycosylated NSP4 is localized to plasma membrane caveolae by a novel raft isolation technique. J. Virol. 2007, 81, 5472–5483. [Google Scholar]
- Matthijnssens, J.; Ciarlet, M.; McDonald, S.M.; Attoui, H.; Bányai, K.; Brister, J.R.; Buesa, J.; Esona, M.D.; Estes, M.K.; Gentsch, J.R.; et al. Uniformity of rotavirus strain nomenclature proposed by the Rotavirus Classification Working Group (RCWG). Arch. Virol. 2011, 156, 1397–1413. [Google Scholar] [CrossRef]
- Kirkwood, C.D.; Coulson, B.; Bishop, R. G3P2 rotaviruses causing diarrhoeal disease in neonates differ in VP4, VP7 y NSP4 sequence from G3P2 strains causing asymptomatic neonatal infection. Arch. Virol. 1996, 141, 1661–1676. [Google Scholar] [CrossRef]
- Pager, C.T.; Alexander, J.J.; Steele, A.D. South African G4P[6] asymptomatic and symptomatic neonatal rotavirus strains differ in their NSP4, VP8*, and VP7 genes. J. Med. Virol. 2000, 62, 208–216. [Google Scholar] [CrossRef]
- Horie, Y.; Masamune, O.; Nakagomi, O. Three major alleles of rotavirus NSP4 proteins identified by sequence analysis. J. Gen. Virol. 1997, 78, 2341–2346. [Google Scholar]
- Ward, R.L.; Mason, B.B.; Bernstein, D.J.; Sander, D.S.; Smith, V.E.; Zandle, G.A.; Rappaport, R.S. Attenuation of a human rotavirus vaccine candidate did not correlate with mutations in the NSP4 protein gene. J. Virol. 1997, 71, 6267–6270. [Google Scholar]
- Lee, C.; Wang, Y.; Kao, C.; Zao, C.; Lee, C.; Chen, H. NSP4 gene analysis of rotaviruses recovered from infected children with and without diarrhea. J. Clin. Microbiol. 2000, 38, 4471–4477. [Google Scholar]
- Parashar, U.D.; Gibson, C.J.; Bresee, J.S.; Glass, R.I. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 2006, 12, 304–306. [Google Scholar] [CrossRef]
- Linhares, A.C.; Velázquez, F.R.; Pérez-Schael, I.; Sáez-Llorens, X.; Abate, H.; Espinoza, F.; López, P.; Macías-Parra, M.; Ortega-Barría, E.; Rivera-Medina, D.M.; et al. Efficacy and safety of an oral live attenuated human rotavirus vaccine against rotavirus gastroenteritis during the first 2 years of life in Latin American infants: Randomised, double-blind controlled study. Lancet 2008, 371, 1181–1189. [Google Scholar] [CrossRef]
- Forster, J.; Guarino, A.; Parez, N.; Moraga, F.; Román, E.; Mory, O.; Tozzi, A.E.; López de Aguileta, A.; Wahn, U.; Graham, C.; et al. Hospital-Based Surveillance to Estimate the Burden of Rotavirus Gastroenteritis Among European Children Younger Than 5 Years of Age. Pediatrics 2009, 123, 393–400. [Google Scholar] [CrossRef]
- Bruijning-Verhagen, P.; Quach, C.; Bonten, M. Nosocomial Rotavirus Infections: A Meta-analysis. Pediatrics 2012, 129, 1011–1019. [Google Scholar] [CrossRef]
- Clemens, J.; Rao, M.; Ahmed, F.; Ward, R.; Huda, S.; Chakraborty, J.; Yunus, M.; Khan, M.R.; Ali, M.; Kay, B.; et al. Breastfeeding and the risk of life-threatening rotavirus diarrhea: Prevention or postponement? Pediatrics 1993, 92, 680–685. [Google Scholar]
- Molyneaux, P. Human immunity to rotavirus. J. Med. Microbiol. 1993, 43, 397–404. [Google Scholar] [CrossRef]
- Raisler, J.; Alexander, C.; O'Campo, P. Breast-feeding and infant illness: A dose-response relationship? Am. J. Public Health 1999, 89, 25–30. [Google Scholar] [CrossRef]
- Ward, R.L.; Bernstein, D.I. Rotarix: A rotavirus vaccine for the world. Clin. Infect. Dis. 2009, 48, 222–228. [Google Scholar] [CrossRef]
- De Palma, O.; Cruz, L.; Ramos, H.; de Baires, A.; Villatoro, N.; Pastor, D.; de Oliveira, L.H.; Kerin, T.; Bowen, M.; Gentsch, J.; et al. Effectiveness of rotavirus vaccination against childhood diarrhoea in El Salvador: Case-control study. BMJ 2010, 340, 2825–2832. [Google Scholar] [CrossRef]
- Munos, M.K.; Fischer, C.L.; Black, R.E. The effect of rotavirus vaccine on diarrhoea mortality. Int. J. Epidemiol. 2010, 39, 56–62. [Google Scholar] [CrossRef]
- Anderson, E.J.; Rupp, A.; Shulman, S.T.; Wang, D.; Zheng, X.; Noskin, G.A. Impact of rotavirus vaccination on hospital-acquired rotavirus gastroenteritis. Pediatrics 2011, 127, 264–270. [Google Scholar] [CrossRef]
- Salinas, B.; Pérez-Schael, I.; Linhares, A.C.; Ruiz-Palacios, G.M.; Guerrero, M.L.; Yarzábal, J.P.; Cervantes, Y.; Costa-Clemens, S.; Damaso, S.; Hardt, K.; De Vos, B. Evaluation of safety, immunogenicity and efficacy of an attenuated rotavirus vaccine, RIX4414: A randomized, placebo-controlled trial in Latin American infants. Pediatr. Infect. Dis. J. 2005, 24, 807–16. [Google Scholar] [CrossRef]
- Ruiz-Palacios, G.M.; Perez-Schael, I.; Velazquez, F.R.; Abate, H.; Breuer, T.; Costa-Clemens, S.; Cheuvart, B.; Espinoza, F.; Gillard, P.; Innis, B.L.; et al. Safety and efficacy of an attenuated vaccine against severe rotavirus gastroenteritis. N. Engl. J. Med. 2006, 354, 11–22. [Google Scholar] [CrossRef]
- Ball, J.M.; Mitchell, D.M.; Gibbons, T.F.; Parr, R. Rotavirus NSP4: A Multifunctional Viral Enterotoxin. Viral Immunol. 2005, 18, 27–40. [Google Scholar] [CrossRef]
- Lorrot, M.; Vasseur, M. How do the rotavirus NSP4 and bacterial enterotoxins lead differently to diarrhea? Virol. J. 2007, 4, 31–37. [Google Scholar] [CrossRef]
- Kudo, S.; Zhou, Y.; Cao, X.; Yamanishi, S.; Nakata, S.; Ushijima, H. Molecular characterization in the VP7, VP4 and NSP4 genes of human rotavirus serotype 4 (G4) isolated in Japan and Kenya. Microbiol. Immunol. 2001, 45, 167–171. [Google Scholar]
- Tavares, T.M.; Brito, W.M.E.D.; Fiaccadori, F.S.; Leal de Freitas, E.R.; Parente, J.A.; Sucasas da Costa, P.S.; Giugliano, L.G.; Andreasi, M.S.A.; Soares, C.M.A.; Cardoso, D.D.P. Molecular characterization of the NSP4 gene of human group A rotavirus samples from the West Central region of Brazil. Mem. Inst. Oswaldo Cruz 2008, 103, 288–294. [Google Scholar] [CrossRef]
- Bányai, K.; Bogdán, A.; Szucs, G.; Arista, S.; De Grazia, S.; Kang, G.; Banerjee, I.; Iturriza-Gómara, M.; Buonavoglia, C.; Martella, V. Assignment of the group A rotavirus NSP4 gene into genotypes using a hemi-nested multiplex PCR assay: A rapid and reproducible assay for strain surveillance studies. J. Med. Microbiol. 2009, 58, 303–311. [Google Scholar] [CrossRef] [Green Version]
- Vizzi, E.; Piñeros, O.; González, G.G.; Zambrano, J.L.; Lurdet, J.E.; Liprandi, F. Genotyping of human rotaviruses circulating among children with diarrhea in Valencia, Venezuela. J. Med. Virol. 2011, 83, 2225–2232. [Google Scholar] [CrossRef]
- Ben Hadj Fredj, M.; Zeller, M.; Fodha, I.; Heylen, E.; Chouikha, A.; Van Ranst, M.; Matthijnssens, J.; Trabelsi, A. Molecular characterization of the NSP4 gene of human group A rotavirus strains circulating in Tunisia from 2006 to 2008. Infect. Genet. Evol. 2012, 12, 997–1004. [Google Scholar] [CrossRef]
- Rahman, M.; Matthijnssens, J.; Yang, X.; Delbeke, T.; Arijs, I.; Taniguchi, K.; Iturriza-Gómara, M.; Iftekharuddin, N.; Azim, T.; Van Ranst, M. Evolutionary history and global spread of the emerging G12 human rotaviruses. J. Virol. 2007, 81, 2382–2390. [Google Scholar] [CrossRef]
- Matthijnssens, J.; Ciarlet, M.; Heiman, E.; Arijs, I.; Delbeke, T.; McDonald, S.M.; Palombo, E.A.; Iturriza-Gómara, M.; Maes, P.; Patton, J.T.; et al. Full genome-based classification of rotaviruses reveals a common origin between human Wa-Like and porcine rotavirus strains and human DS-1-like and bovine rotavirus strains. J. Virol. 2008, 82, 3204–3219. [Google Scholar] [CrossRef]
- Benati, F.J.; Maranhao, A.G.; Lima, R.S.; da Silva, RC.; Santos, N. Multiple-gene characterization of rotavirus strains: Evidence of genetic linkage among the VP7-, VP4-, VP6-, and NSP4-encoding genes. J. Med. Virol. 2010, 82, 1797–1802. [Google Scholar] [CrossRef]
- Khamrin, P.; Maneekarn, N.; Malasao, R.; Nguyen, T.A.; Ishida, S.; Okitsu, S.; Ushijima, H. Genotypic linkages of VP4, VP6, VP7, NSP4, NSP5 genes of rotaviruses circulating among children with acute gastroenteritis in Thail. Infect. Genet. Evol. 2010, 10, 467–472. [Google Scholar]
- Ghosh, S.; Gatheru, Z.; Nyangao, J.; Adachi, N.; Urushibara, N.; Kobayashi, N. Full genomic analysis of a simian SA11-like G3P[2] rotavirus strain isolated from an asymptomatic infant: Identification of novel VP1, VP6 and NSP4 genotypes. Infect. Genet. Evol. 2011, 11, 57–63. [Google Scholar] [CrossRef]
- Kirkwood, C.D.; Gentsch, J.R.; Glass, R.I. Sequence analysis of the NSP4 gene from human rotavirus strains isolated in the United States. Virus Genes 1999, 19, 113–122. [Google Scholar] [CrossRef]
- Araujo, I.T.; Heinemann, M.B.; Mascarenhas, J.D.P.; Santos Assis, R.M.; Fialho, A.M.; Leite, J.P.G. Molecular analysis of the NSP4 and VP6 genes of rotavirus strains recovered from hospitalized children in Rio de Janeiro, Brazil. J. Med. Microbiol. 2007, 56, 854–859. [Google Scholar] [CrossRef]
- Betts, M.J.; Russell, R.B. Amino Acid properties and Consequences of Substitutions. In Bioinformatics for Geneticists; Barnes, M.R., Gray, I.C., Eds.; Wiley: New York, New York, USA, 2003. [Google Scholar]
- Bowman, G.D.; Nodelman, I.M.; Levy, O.; Lin, S.L.; Tian, P.; Zamb, T.J.; Udem, S.A.; Venkataraghavan, B.; Schutt, C.E. Crystal structure of the oligomerization domain of NSP4 from rotavirus reveals a core metal-binding site. J. Mol. Biol. 2000, 304, 861–871. [Google Scholar] [CrossRef]
- Seo, N.; Zeng, CQ-Y, Hyser; Utama, B.; Crawford, S.E.; Kim, K.J.; Höök, M.; Estes, M.K. Integrins α1β1 and α2β1 are receptors for the rotavirus enterotoxin. Proc. Natl. Acad. Sci. USA 2008, 105, 8811–8818. [Google Scholar]
- Ruuska, T.; Vesikari, T. Rotavirus disease in Finnish children: use of numerical scoresfor clinical severity of diarrhoeal episodes. Scand. J. Infect. Dis. 1991, 22, 259–267. [Google Scholar] [CrossRef]
- Verly, E.; Cohen, J. Demostration of size variation of RNA segments between different isolates of calf rotavirus. J. Gen. Virol. 1977, 35, 583–586. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
González-Ochoa, G.; Menchaca, G.E.; Hernández, C.E.; Rodríguez, C.; Tamez, R.S.; Contreras, J.F. Mutation Distribution in the NSP4 Protein in Rotaviruses Isolated from Mexican Children with Moderate to Severe Gastroenteritis. Viruses 2013, 5, 792-805. https://doi.org/10.3390/v5030792
González-Ochoa G, Menchaca GE, Hernández CE, Rodríguez C, Tamez RS, Contreras JF. Mutation Distribution in the NSP4 Protein in Rotaviruses Isolated from Mexican Children with Moderate to Severe Gastroenteritis. Viruses. 2013; 5(3):792-805. https://doi.org/10.3390/v5030792
Chicago/Turabian StyleGonzález-Ochoa, Guadalupe, Griselda E. Menchaca, Carlos E. Hernández, Cristina Rodríguez, Reyes S. Tamez, and Juan F. Contreras. 2013. "Mutation Distribution in the NSP4 Protein in Rotaviruses Isolated from Mexican Children with Moderate to Severe Gastroenteritis" Viruses 5, no. 3: 792-805. https://doi.org/10.3390/v5030792




