A Polytropic Caprine Arthritis Encephalitis Virus Promoter Isolated from Multiple Tissues from a Sheep with Multisystemic Lentivirus-Associated Inflammatory Disease
Abstract
:1. Introduction
2. Results
2.1. Pathology and Immunohistochemistry

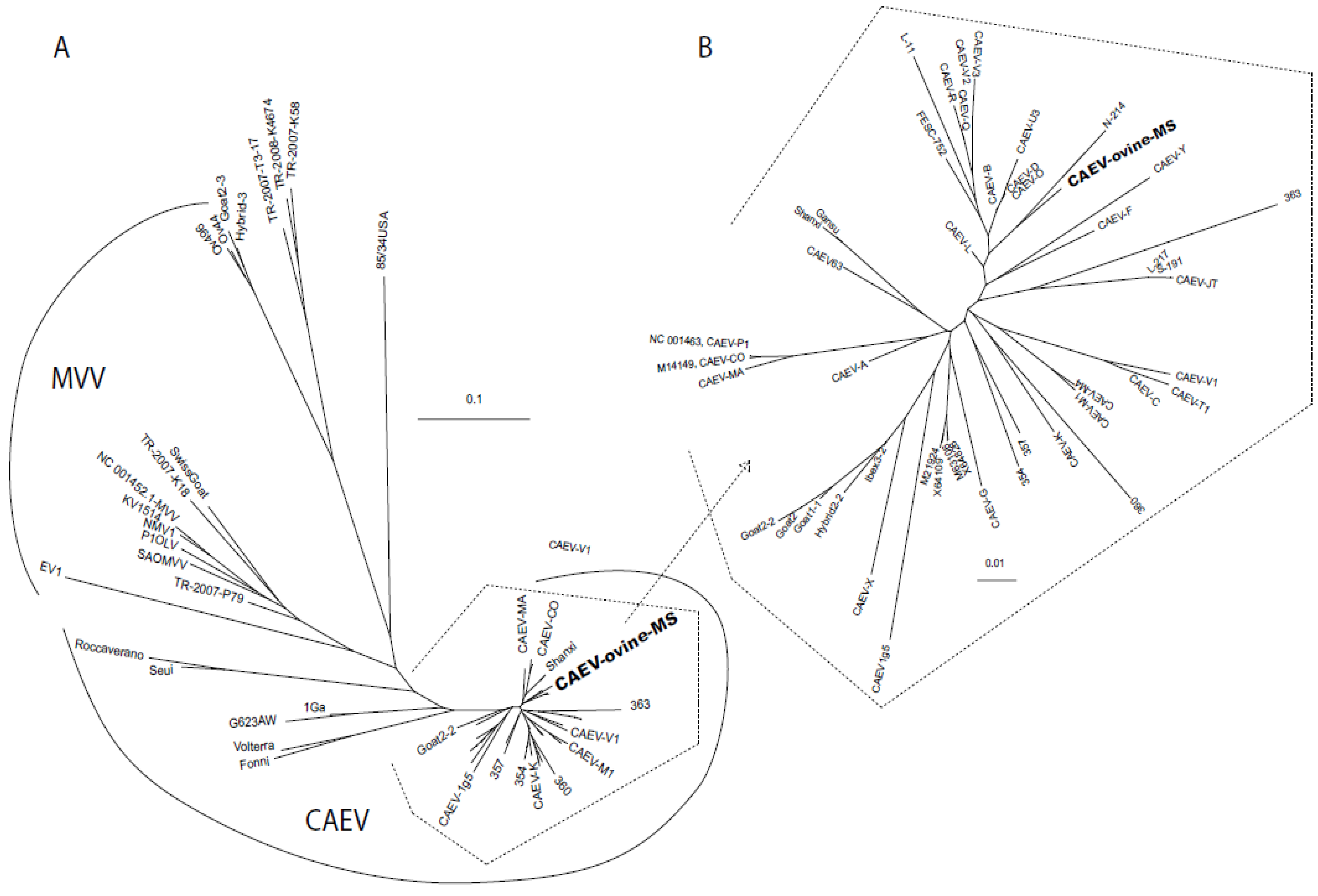
2.2. Sequencing and Phylogenetic Analysis
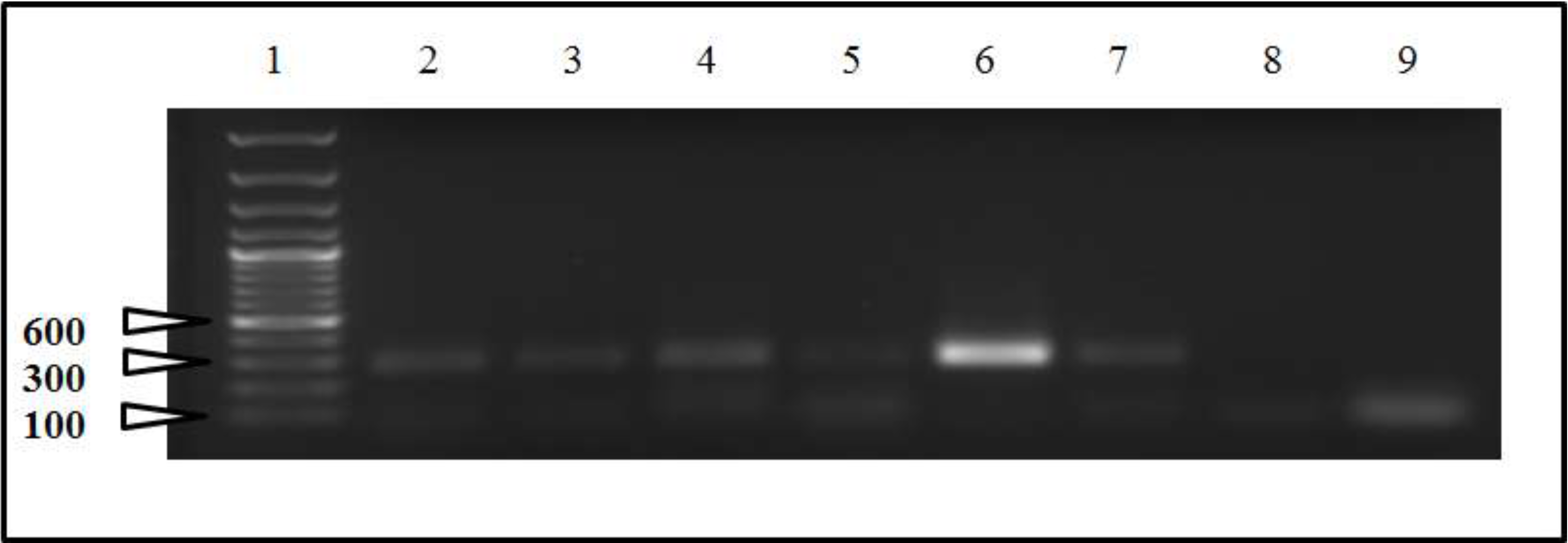
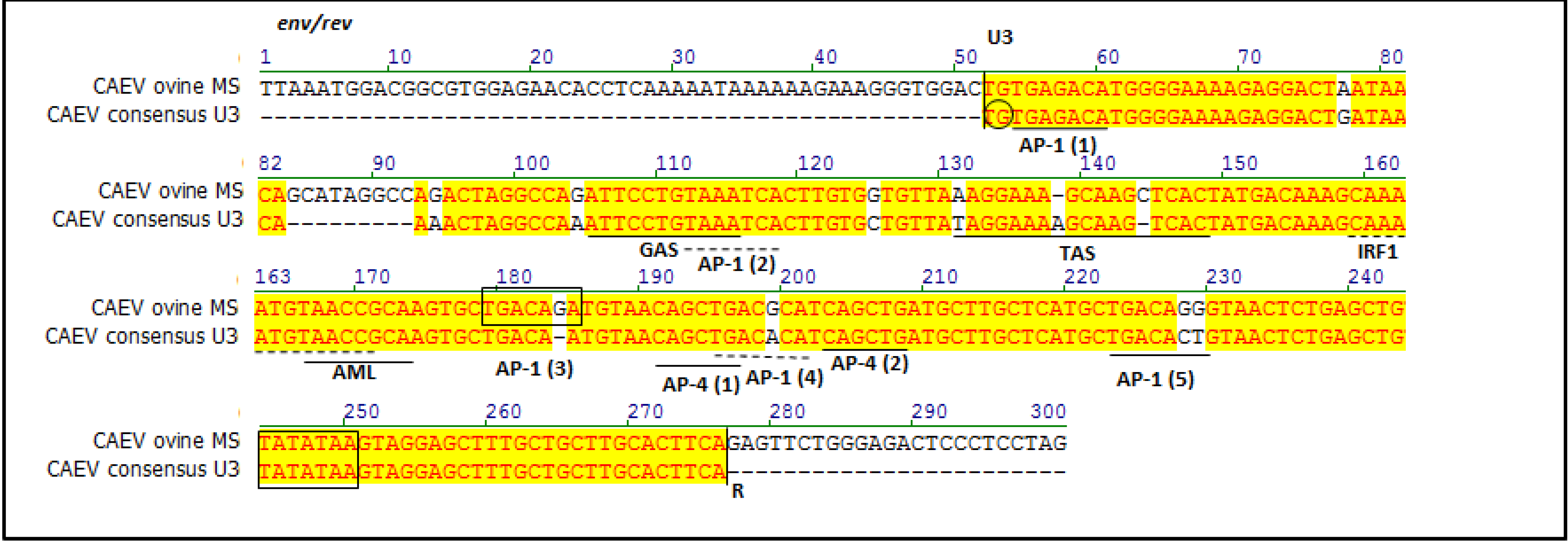
3. Discussion
4. Materials and Methods
4.1. Signalment and Pathology
4.2. Immunohistochemistry and Serology
4.3. Molecular Techniques
4.4. Phylogenetic and Transcription Factor Analyses
5. Conclusion
Conflict of Interest
Acknowledgements
References and Notes
- Benavides, J.; Garcia-Pariente, C.; Fuertes, M.; Ferreras, M.C.; Garcia-Marin, J.F.; Juste, R.A.; Perez, V. Maedi-visna: The meningoencephalitis in naturally occurring cases. J. Comp. Pathol. 2009, 140, 1–11. [Google Scholar] [CrossRef]
- Fields Virology, 4th ed.; Knipe, D.M.; Howley, P.M.; Griffin, D.E.; Lamb, R.A.; Martin, A.M.; Roizman, B.; Straus, E.S. (Eds.) Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2001.
- Maxie, M.G.; Jubb, K.V.F. Pathology of Domestic Animals, 5th ed.; Elsevier Saunders: Edinburgh ; New York, NY, USA, 2007. [Google Scholar]
- Zink, M.C.; Yager, J.A.; Myers, J.D. Pathogenesis of caprine arthritis encephalitis virus. Cellular localization of viral transcripts in tissues of infected goats. Am. J. Pathol. 1990, 136, 843–854. [Google Scholar]
- Ryan, S.; Tiley, L.; McConnell, I.; Blacklaws, B. Infection of dendritic cells by the Maedi-Visna lentivirus. J. Virol. 2000, 74, 10096–10103. [Google Scholar] [CrossRef]
- Kennedy-Stoskopf, S.; Zink, M.C.; Jolly, P.E.; Narayan, O. Lentivirus-induced arthritis. Chronic disease caused by a covert pathogen. Rheum. Dis. Clin. North. Am. 1987, 13, 235–247. [Google Scholar]
- Benavides, J.; Fuertes, M.; Garcia-Pariente, C.; Otaola, J.; Delgado, L.; Giraldez, J.; Garcia Marin, J.F.; Carmen Ferreras, M.; Perez, V. Impact of maedi-visna in intensively managed dairy sheep. Vet. J. 2013. [Google Scholar] [CrossRef]
- Rachid, A.; Croise, B.; Russo, P.; Vignoni, M.; Lacerenza, D.; Rosati, S.; Kuzmak, J.; Valas, S. Diverse host-virus interactions following caprine arthritis-encephalitis virus infection in sheep and goats. J. Gen. Virol. 2013, 94, 634–642. [Google Scholar] [CrossRef]
- Cork, L.C. Differential diagnosis of viral leukoencephalomyelitis of goats. J. Am. Vet. Med. Assoc. 1976, 169, 1303–1306. [Google Scholar]
- Cork, L.C.; Davis, W.C. Ultrastructural features of viral leukoencephalomyelitis of goats. Lab. Invest. 1975, 32, 359–365. [Google Scholar]
- Cork, L.C.; Hadlow, W.J.; Crawford, T.B.; Gorham, J.R.; Piper, R.C. Infectious leukoencephalomyelitis of young goats. J. Infect. Dis. 1974, 129, 134–141. [Google Scholar] [CrossRef]
- Cork, L.C.; Hadlow, W.J.; Gorham, J.R.; Piper, R.C.; Crawford, T.B. Pathology of viral leukoencephalomyelitis of goats. Acta Neuropathol. 1974, 29, 281–292. [Google Scholar] [CrossRef]
- Murphy, B.; McElliott, V.; Vapniarsky, N.; Oliver, A.; Rowe, J. Tissue tropism and promoter sequence variation in caprine arthritis encephalitis virus infected goats. Virus Res. 2010, 151, 177–184. [Google Scholar] [CrossRef]
- Agnarsdottir, G.; Thorsteinsdottir, H.; Oskarsson, T.; Matthiasdottir, S.; Haflidadottir, B.S.; Andresson, O.S.; Andresdottir, V. The long terminal repeat is a determinant of cell tropism of maedi-visna virus. J. Gen. Virol. 2000, 81, 1901–1905. [Google Scholar]
- Barros, S.C.; Andresdottir, V.; Fevereiro, M. Cellular specificity and replication rate of Maedi Visna virus in vitro can be controlled by LTR sequences. Arch. Virol. 2005, 150, 201–213. [Google Scholar] [CrossRef]
- Ross, H.L.; Gartner, S.; McArthur, J.C.; Corboy, J.R.; McAllister, J.J.; Millhouse, S.; Wigdahl, B. HIV-1 LTR C/EBP binding site sequence configurations preferentially encountered in brain lead to enhanced C/EBP factor binding and increased LTR-specific activity. J. Neurovirol. 2001, 7, 235–249. [Google Scholar] [CrossRef]
- Palmarini, M.; Datta, S.; Omid, R.; Murgia, C.; Fan, H. The long terminal repeat of Jaagsiekte sheep retrovirus is preferentially active in differentiated epithelial cells of the lungs. J. Virol. 2000, 74, 5776–5787. [Google Scholar] [CrossRef]
- Valas, S.; Benoit, C.; Baudry, C.; Perrin, G.; Mamoun, R.Z. Variability and immunogenicity of caprine arthritis-encephalitis virus surface glycoprotein. J. Virol. 2000, 74, 6178–6185. [Google Scholar] [CrossRef]
- Murphy, B.; Hillman, C.; Castillo, D.; Vapniarsky, N.; Rowe, J. The presence or absence of the gamma-activated site determines IFN gamma-mediated transcriptional activation in CAEV promoters cloned from the mammary gland and joint synovium of a single CAEV-infected goat. Virus Res. 2012, 163, 537–545. [Google Scholar] [CrossRef]
- Knowles, D.P., Jr.; Cheevers, W.P.; McGuire, T.C.; Brassfield, A.L.; Harwood, W.G.; Stem, T.A. Structure and genetic variability of envelope glycoproteins of two antigenic variants of caprine arthritis-encephalitis lentivirus. J. Virol. 1991, 65, 5744–5750. [Google Scholar]
- Gomez-Lucia, E.; Rowe, J.; Collar, C.; Murphy, B. Diversity of caprine arthritis-encephalitis virus promoters isolated from goat milk and passaged in vitro. Vet. J. 2013, 196, 431–438. [Google Scholar] [CrossRef]
- Transcription Factor Search. Available online: http://www.cbrc.jp/research/db/TFSEARCH.html/ (accessed on 22 June 2013).
- Herrmann, L.M.; Cheevers, W.P.; Marshall, K.L.; McGuire, T.C.; Hutton, M.M.; Lewis, G.S.; Knowles, D.P. Detection of serum antibodies to ovine progressive pneumonia virus in sheep by using a caprine arthritis-encephalitis virus competitive-inhibition enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 2003, 10, 862–865. [Google Scholar]
- Pescador, C.A.; Corbellini, L.G.; Oliveira, E.C.; Raymundo, D.L.; Driemeier, D. Histopathological and immunohistochemical aspects of Neospora caninum diagnosis in bovine aborted fetuses. Vet. Parasitol. 2007, 150, 159–163. [Google Scholar] [CrossRef]
- Hamir, A.N.; Moser, G.; Galligan, D.T.; Davis, S.W.; Granstrom, D.E.; Dubey, J.P. Immunohistochemical study to demonstrate Sarcocystis neurona in equine protozoal myeloencephalitis. J. Vet. Diagn. Invest. 1993, 5, 418–422. [Google Scholar] [CrossRef]
- Monleon, E.; Monzon, M.; Hortells, P.; Bolea, R.; Acin, C.; Vargas, F.; Badiola, J.J. Approaches to Scrapie diagnosis by applying immunohistochemistry and rapid tests on central nervous and lymphoreticular systems. J. Virol. Methods 2005, 125, 165–171. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Page, R.D. TreeView: An application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 1996, 12, 357–358. [Google Scholar]
Supplementary Files
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Adedeji, A.O.; Barr, B.; Gomez-Lucia, E.; Murphy, B. A Polytropic Caprine Arthritis Encephalitis Virus Promoter Isolated from Multiple Tissues from a Sheep with Multisystemic Lentivirus-Associated Inflammatory Disease. Viruses 2013, 5, 2005-2018. https://doi.org/10.3390/v5082005
Adedeji AO, Barr B, Gomez-Lucia E, Murphy B. A Polytropic Caprine Arthritis Encephalitis Virus Promoter Isolated from Multiple Tissues from a Sheep with Multisystemic Lentivirus-Associated Inflammatory Disease. Viruses. 2013; 5(8):2005-2018. https://doi.org/10.3390/v5082005
Chicago/Turabian StyleAdedeji, Adeyemi O., Bradd Barr, Esperanza Gomez-Lucia, and Brian Murphy. 2013. "A Polytropic Caprine Arthritis Encephalitis Virus Promoter Isolated from Multiple Tissues from a Sheep with Multisystemic Lentivirus-Associated Inflammatory Disease" Viruses 5, no. 8: 2005-2018. https://doi.org/10.3390/v5082005



