Non-Simian Foamy Viruses: Molecular Virology, Tropism and Prevalence and Zoonotic/Interspecies Transmission
Abstract
:1. Defining Statement
2. Classification of a Complex Group of Viruses
2.1. Natural History and Genome Organization
| Clade/isolate | Host | References |
|---|---|---|
| SFV (incl. PFV) | simians, apes and humans | [8,10,22,23,24,25,26] |
| FFV | cats (domestic and wild) | [27,28,29] |
| BFV | cattle | [30,31,32] |
| EFV | horse | [33] |
| RaFV-1 | bat | [14] |
| CoeEFV | coelacanth (endogeneous) | [13] |
| SloEFV | sloth (endogenous) | [12] |
| PSFVaye | Daubentonia madagascariensis (aye-aye) | [15] |
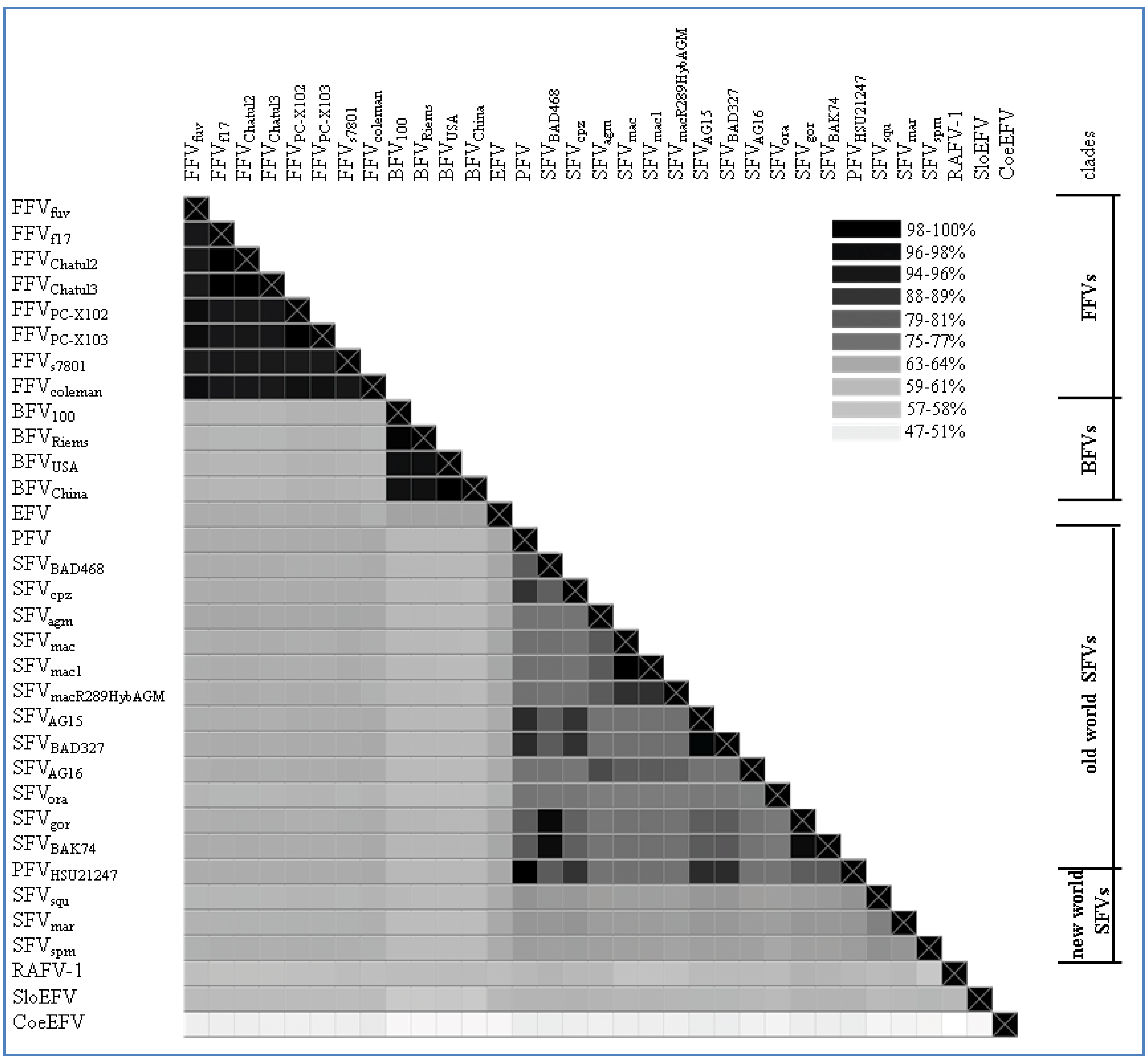
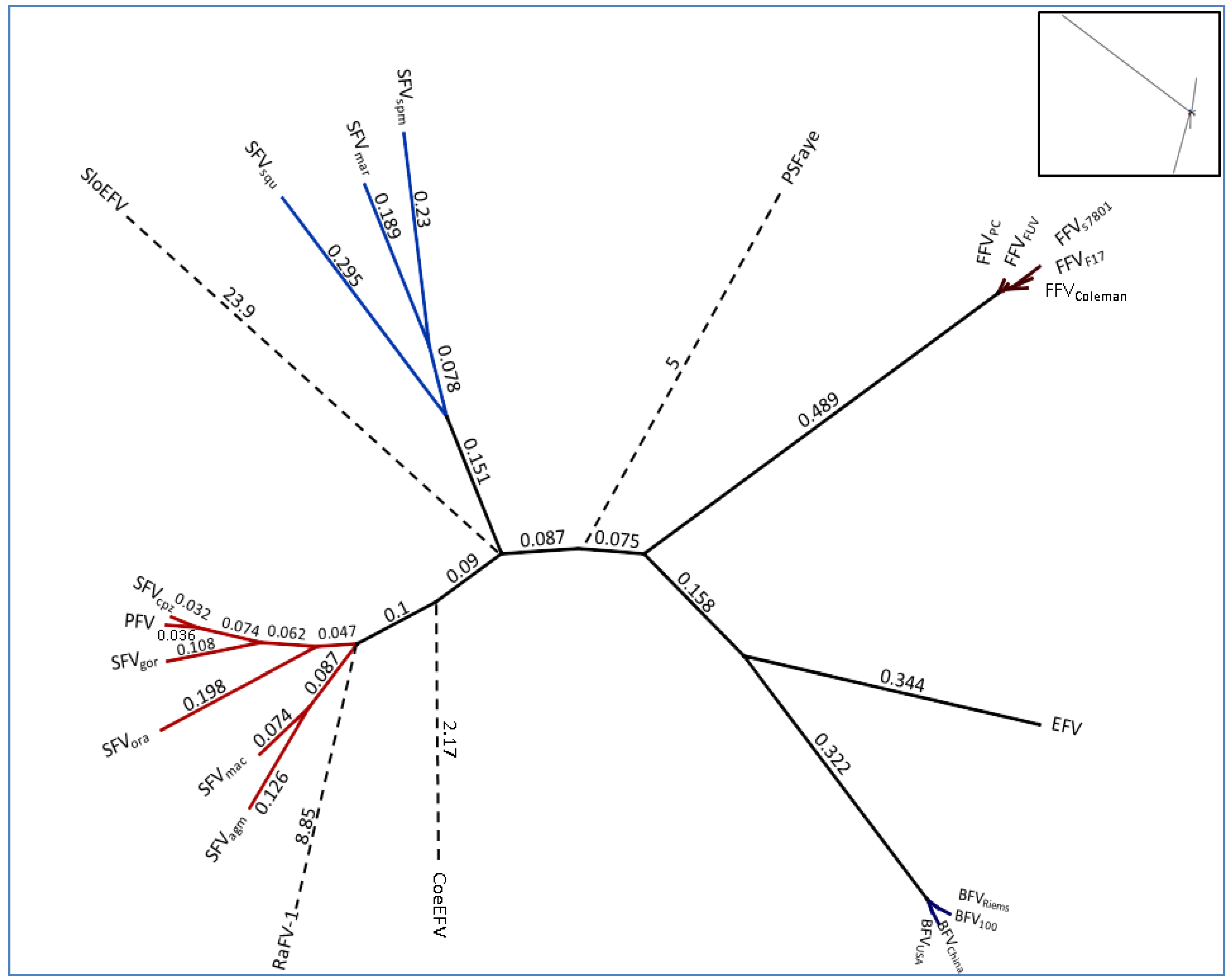
2.2. Overall Genome-Wide Comparison of Animal FVs
2.3. Feline Foamy Virus Clade
| Acronym | Description | Virus clade/isolates | Accession number |
|---|---|---|---|
| BFV100 | Bovine Foamy Virus BFV100 isolate, Poland | Bovine foamy virus | JX307861.1 |
| BFVChina | Bovine Foamy Virus Geng isolate, China | Bovine foamy virus | AY134750 |
| BFVRiems | Bovine Foamy Virus Riems isolate, Germany | Bovine foamy virus | JX307862.1 |
| BFVUSA | Bovine Foamy Virus Casey isolate, USA | Bovine foamy virus | U94514 |
| EFV | Equine Foamy Virus isolate | Equine Foamy Virus | AF201902 |
| FFVFUV | Feline Foamy Virus FUV ioslate, Germany | Feline foamy Virus | Y08851 |
| FFVPC | Feline Foamy Virus Puma C. isolate, USA | Feline foamy Virus | KC292054 |
| FFVF17 | Feline Foamy Virus F17 isolate, USA | Feline foamy Virus | NC_001871.1 |
| FFVColeman | Feline Foamy Virus Coleman isolate, USA | Feline foamy Virus | AB052797 |
| FFVS7801 | Feline Foamy Virus S7801 isolate, Japan | Feline foamy Virus | AB052796 |
| SFVagm | Simian foamy virus 3, Vervet monkey | Simian foamy Virus | NC_010820 |
| SFVcpz | Simian Foamy Virus, Chimpanzee | Simian foamy Virus | U04327 |
| SFVgor | Simian Foamy Virus, Gorilla | Simian foamy Virus | HM245790 |
| SFVmac | Simian Foamy Virus 1, Macaque | Simian foamy Virus | X54482 |
| SFVora | Simian Foamy Virus, Orang Utan | Simian foamy Virus | AJ544579 |
| PFV | Human Foamy Virus isolate | Simian foamy Virus | U21247 |
| SFVspm | Simian Foamy Virus, Spider monkey | Simian foamy Virus | EU010385.1 |
| SFVmar | Simian Foamy Virus, Marmoset monkey | Simian foamy Virus | GU356395 |
| SFVsqu | Simian Foamy Virus, Squirrel monkey | Simian foamy Virus | GU356394 |
| SFVgor | Simian Foamy Virus, Gorilla | Simian foamy Virus | HM245790 |
| SloEFV | Endogenous Foamy Virus, Sloth | Sloth Foamy Virus | [12] |
| CoeEFV | Endogenous Foamy Virus, Coelacanth | Coelacanth Foamy Virus | [13] |
| RaFV-1 | Exo- or endogenous Foamy Virus, Bat | Bat Foamy Virus | [14] |
2.4. Ungulate Foamy Virus Clade
2.5. Simian Foamy Virus Clade
2.6. “Exotic” Foamy Viruses
2.7. Conservation of FV Specific Traits
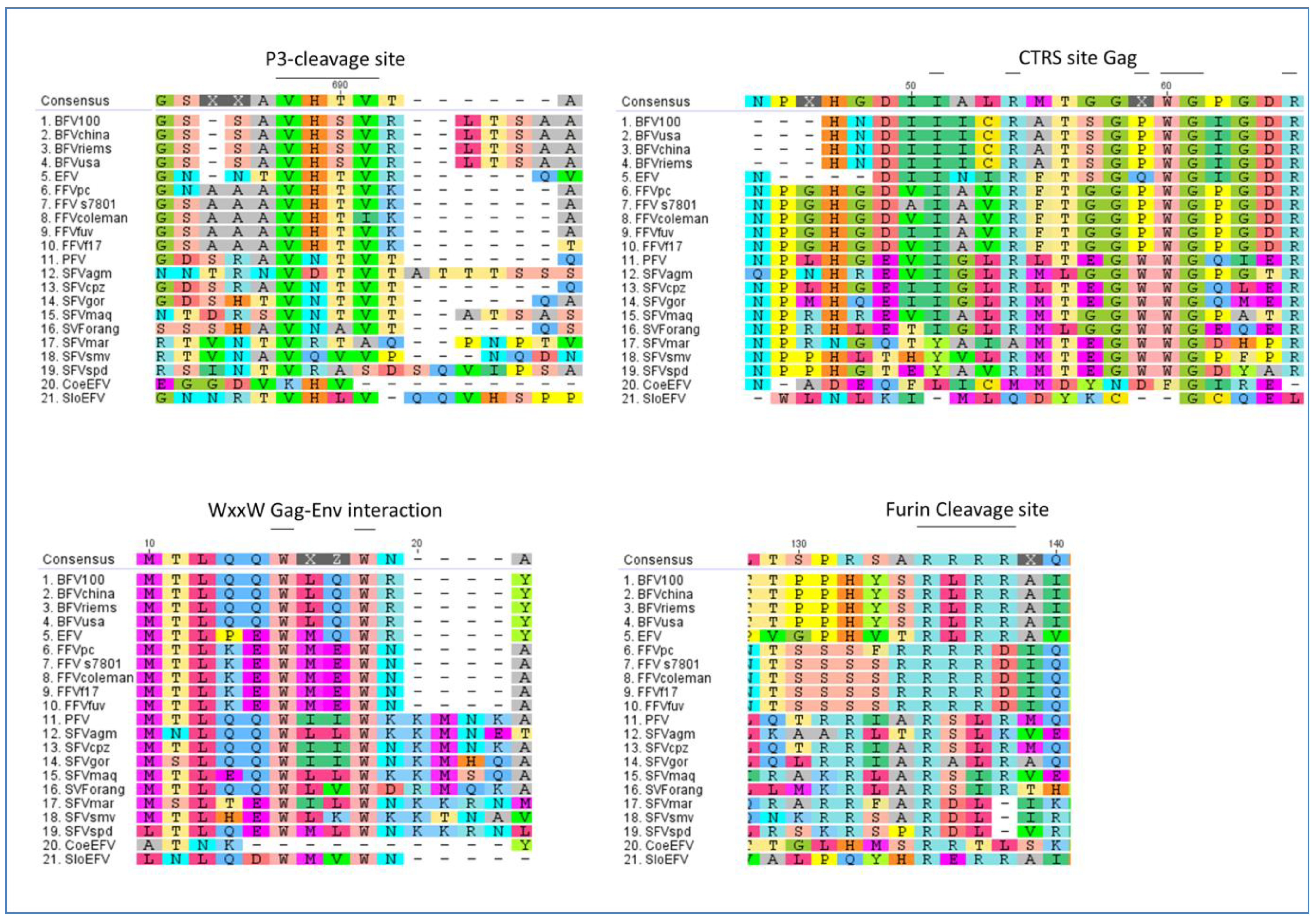
3. Molecular Biology of FV
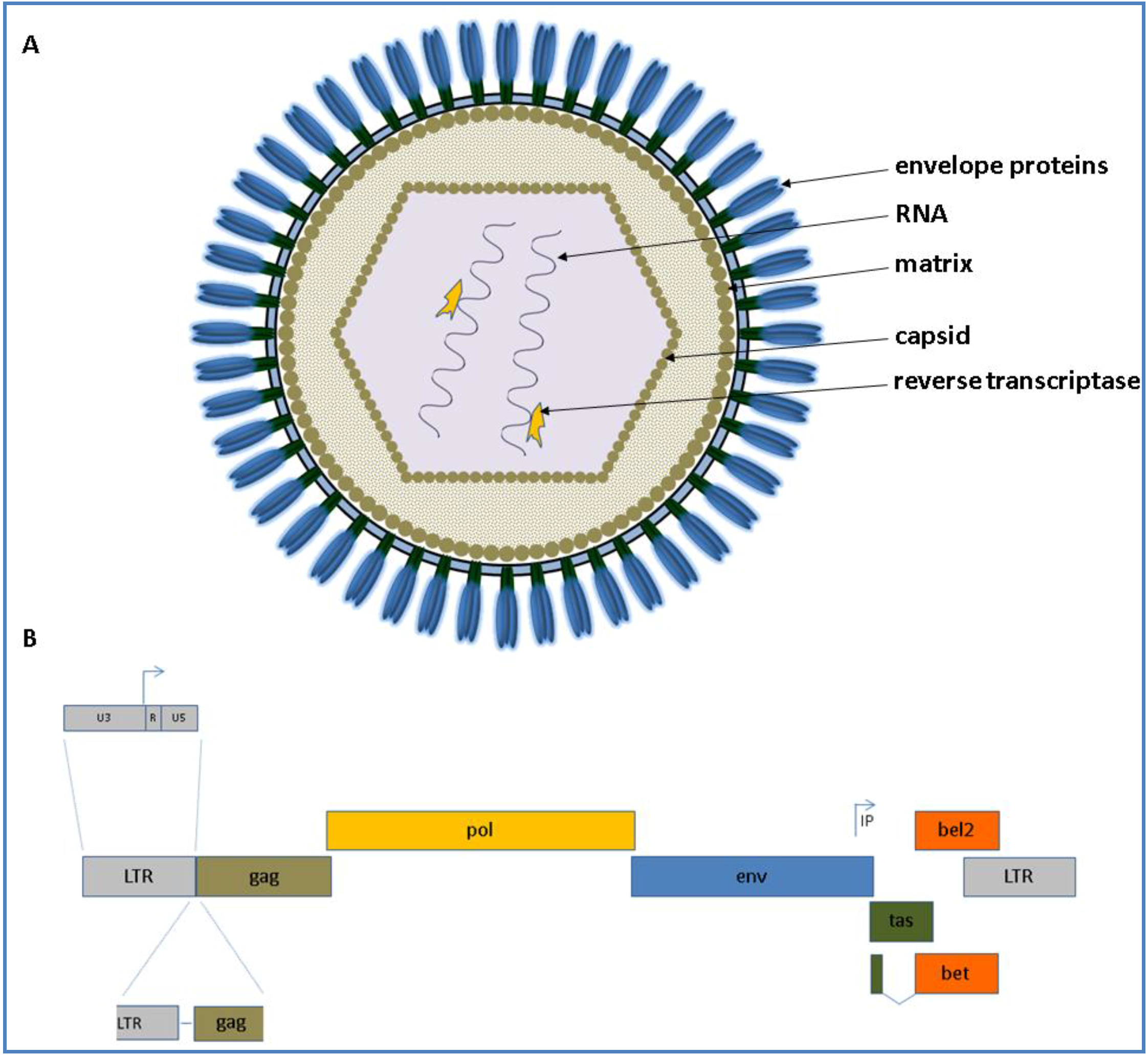
3.1. Virus Structure and Morphogenesis
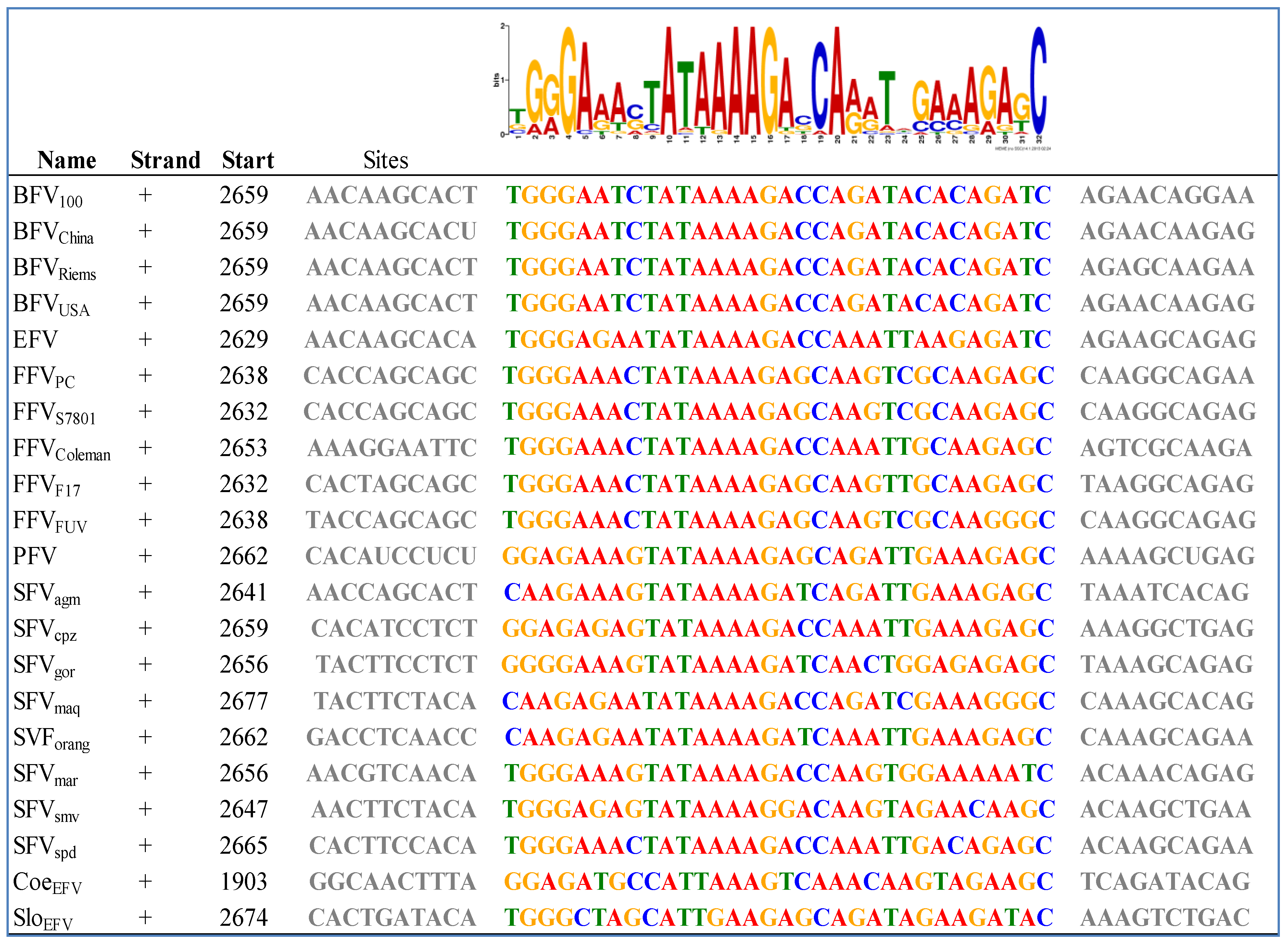
3.2. Foamy Viral Transcription
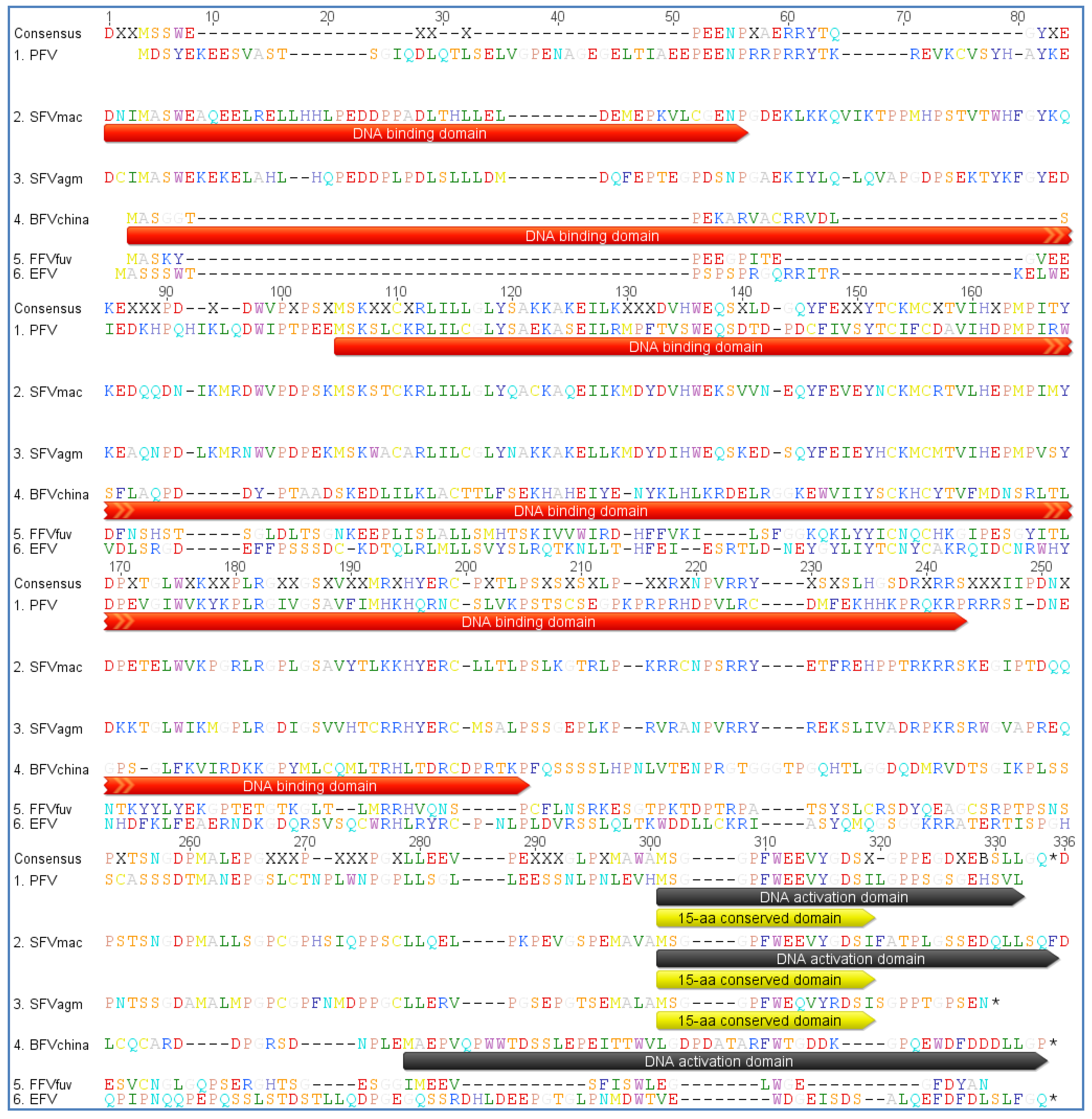

3.3. Host Restriction
4. Biology in the Host
4.1. FV Tropism
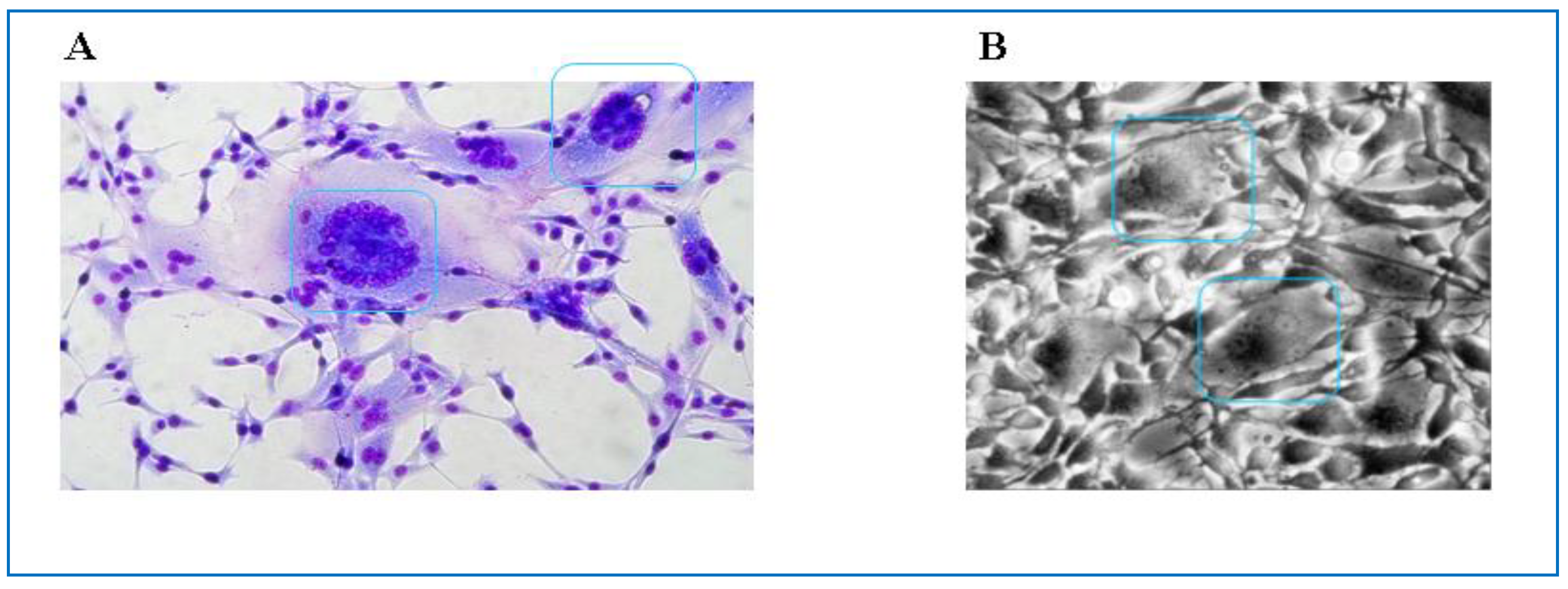
4.2. FVs Prevalence
4.3. Viral Transmission
4.4. Interspecies Transmission
4.5. Zoonotic Potential
5. FV Diagnostic Methods
5.1. Immunological Methods
5.2. Molecular Methods
6. Perspectives for Future Research
Acknowledgments
Conflicts of Interest
References and Notes
- Saib, A. Non-primate foamy viruses. Curr. Top. Microbiol. Immunol. 2003, 277, 197–211. [Google Scholar] [CrossRef]
- Delelis, O.; Lehmann-Che, J.; Saib, A. Foamy viruses—A world apart. Curr. Opin. Microbiol. 2004, 7, 400–406. [Google Scholar] [CrossRef]
- Murray, S.M.; Linial, M.L. Foamy virus infection in primates. J. Med. Primatol. 2006, 35, 225–235. [Google Scholar] [CrossRef]
- Rethwilm, A. Molecular biology of foamy viruses. Med. Microbiol. Immunol. 2010, 199, 197–207. [Google Scholar] [CrossRef]
- Meiering, C.D.; Linial, M.L. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 2001, 14, 165–176. [Google Scholar] [CrossRef]
- Enders, J.F.; Peebles, T.C. Propagation in tissue culture of cytopathogenic agents from patients with measles. Proc. Soc. Exp. Biol. Med. 1954, 86, 277–286. [Google Scholar] [CrossRef]
- Achong, B.G.; Mansell, P.W.; Epstein, M.A.; Clifford, P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J. Natl. Canc. Inst. 1971, 46, 299–307. [Google Scholar]
- Herchenroder, O.; Renne, R.; Loncar, D.; Cobb, E.K.; Murthy, K.K.; Schneider, J.; Mergia, A.; Luciw, P.A. Isolation, cloning, and sequencing of simian foamy viruses from chimpanzees (sfvcpz): High homology to human foamy virus (hfv). Virology 1994, 201, 187–199. [Google Scholar] [CrossRef]
- Switzer, W.M.; Bhullar, V.; Shanmugam, V.; Cong, M.E.; Parekh, B.; Lerche, N.W.; Yee, J.L.; Ely, J.J.; Boneva, R.; Chapman, L.E.; et al. Frequent simian foamy virus infection in persons occupationally exposed to nonhuman primates. J. Virol. 2004, 78, 2780–2789. [Google Scholar] [CrossRef]
- Schulze, A.; Lemey, P.; Schubert, J.; McClure, M.O.; Rethwilm, A.; Bodem, J. Complete nucleotide sequence and evolutionary analysis of a gorilla foamy virus. J. Gen. Virol. 2011, 92, 582–586. [Google Scholar] [CrossRef]
- Switzer, W.M.; Salemi, M.; Shanmugam, V.; Gao, F.; Cong, M.E.; Kuiken, C.; Bhullar, V.; Beer, B.E.; Vallet, D.; Gautier-Hion, A.; et al. Ancient co-speciation of simian foamy viruses and primates. Nature 2005, 434, 376–380. [Google Scholar] [CrossRef]
- Katzourakis, A.; Gifford, R.J.; Tristem, M.; Gilbert, M.T.; Pybus, O.G. Macroevolution of complex retroviruses. Science 2009, 325, 1512. [Google Scholar] [CrossRef]
- Han, G.Z.; Worobey, M. An endogenous foamy-like viral element in the coelacanth genome. PLoS Pathog. 2012, 8, e1002790. [Google Scholar] [CrossRef]
- Wu, Z.; Ren, X.; Yang, L.; Hu, Y.; Yang, J.; He, G.; Zhang, J.; Dong, J.; Sun, L.; Du, J.; et al. Virome analysis for identification of novel mammalian viruses in bat species from chinese provinces. J. Virol. 2012, 86, 10999–11012. [Google Scholar] [CrossRef]
- Han, G.Z.; Worobey, M. An endogenous foamy virus in the aye-aye (daubentonia madagascariensis). J. Virol. 2012, 86, 7696–7698. [Google Scholar] [CrossRef]
- Linial, M. Why aren't foamy viruses pathogenic? Trends Microbiol. 2000, 8, 284–289. [Google Scholar] [CrossRef]
- Wilk, T.; Geiselhart, V.; Frech, M.; Fuller, S.D.; Flugel, R.M.; Lochelt, M. Specific interaction of a novel foamy virus env leader protein with the n-terminal gag domain. J. Virol. 2001, 75, 7995–8007. [Google Scholar] [CrossRef]
- Kennedy-Stoskopf, S.; Stoskopf, M.K.; Eckhaus, M.A.; Strandberg, J.D. Isolation of a retrovirus and a herpesvirus from a captive california sea lion. J. Wildl. Dis. 1986, 22, 156–164. [Google Scholar]
- Flanagan, M. Isolation of a spumavirus from a sheep. Aust. Vet. J. 1992, 69. [Google Scholar]
- Materniak, M.; Kuzmak, J.; Olech, M. Seroreactivity to bovine foamy virus antigens in wild ruminants—Evidence for new reservoirs of foamy viruses. In Proceedings of XIII DIAGMOL Conference, Warsaw, Poland, 24 November 2012.
- Hruska, J.F.; Takemoto, K.K. Biochemical properties of a hamster syncytium-forming ("foamy") virus. J. Natl. Canc. Inst. 1975, 54, 601–605. [Google Scholar]
- McClure, M.O.; Bieniasz, P.D.; Schulz, T.F.; Chrystie, I.L.; Simpson, G.; Aguzzi, A.; Hoad, J.G.; Cunningham, A.; Kirkwood, J.; Weiss, R.A. Isolation of a new foamy retrovirus from orangutans. J. Virol. 1994, 68, 7124–7130. [Google Scholar]
- Pacheco, B.; Finzi, A.; McGee-Estrada, K.; Sodroski, J. Species-specific inhibition of foamy viruses from south american monkeys by new world monkey trim5{alpha} proteins. J. Virol. 2010, 84, 4095–4099. [Google Scholar] [CrossRef]
- Renne, R.; Friedl, E.; Schweizer, M.; Fleps, U.; Turek, R.; Neumann-Haefelin, D. Genomic organization and expression of simian foamy virus type 3 (sfv-3). Virology 1992, 186, 597–608. [Google Scholar] [CrossRef]
- Thumer, L.; Rethwilm, A.; Holmes, E.C.; Bodem, J. The complete nucleotide sequence of a new world simian foamy virus. Virology 2007, 369, 191–197. [Google Scholar] [CrossRef]
- Verschoor, E.J.; Langenhuijzen, S.; van den Engel, S.; Niphuis, H.; Warren, K.S.; Heeney, J.L. Structural and evolutionary analysis of an orangutan foamy virus. J. Virol. 2003, 77, 8584–8587. [Google Scholar] [CrossRef]
- Bodem, J.; Lochelt, M.; Winkler, I.; Flower, R.P.; Delius, H.; Flugel, R.M. Characterization of the spliced pol transcript of feline foamy virus: The splice acceptor site of the pol transcript is located in gag of foamy viruses. J. Virol. 1996, 70, 9024–9027. [Google Scholar]
- Winkler, I.; Bodem, J.; Haas, L.; Zemba, M.; Delius, H.; Flower, R.; Flugel, R.M.; Lochelt, M. Characterization of the genome of feline foamy virus and its proteins shows distinct features different from those of primate spumaviruses. J. Virol. 1997, 71, 6727–6741. [Google Scholar]
- Kehl, T.; Bleiholder, A.; Rossmann, F.; Rupp, S.; Lei, J.; Lee, J.; Boyce, W.M.; Vickers, W.; Crooks, K.; VandeWoude, S.; Loechelt, M. Complete genome sequences of two novel puma concolor foamy viruses from California. Genome Announc. 2013, 1, e00201-12. [Google Scholar]
- Materniak, M.; Bicka, L.; Kuzmak, J. Isolation and partial characterization of bovine foamy virus from polish cattle. Pol. J. Vet. Sci. 2006, 9, 207–211. [Google Scholar]
- Hechler, T.; Materniak, M.; Kehl, T.; Kuzmak, J.; Lochelt, M. Complete genome sequences of two novel european clade bovine foamy viruses from germany and poland. J. Virol. 2012, 86, 10905–10906. [Google Scholar] [CrossRef]
- Holzschu, D.L.; Delaney, M.A.; Renshaw, R.W.; Casey, J.W. The nucleotide sequence and spliced pol mrna levels of the nonprimate spumavirus bovine foamy virus. J. Virol. 1998, 72, 2177–2182. [Google Scholar]
- Tobaly-Tapiero, J.; Bittoun, P.; Neves, M.; Guillemin, M.C.; Lecellier, C.H.; Puvion-Dutilleul, F.; Gicquel, B.; Zientara, S.; Giron, M.L.; de The, H.; et al. Isolation and characterization of an equine foamy virus. J. Virol. 2000, 74, 4064–4073. [Google Scholar] [CrossRef]
- Phung, H.T.; Ikeda, Y.; Miyazawa, T.; Nakamura, K.; Mochizuki, M.; Izumiya, Y.; Sato, E.; Nishimura, Y.; Tohya, Y.; Takahashi, E.; et al. Genetic analyses of feline foamy virus isolates from domestic and wild feline species in geographically distinct areas. Virus Res. 2001, 76, 171–181. [Google Scholar] [CrossRef]
- Flower, R.L.; Wilcox, G.E.; Cook, R.D.; Ellis, T.M. Detection and prevalence of serotypes of feline syncytial spumaviruses. Arch. Virol. 1985, 83, 53–63. [Google Scholar] [CrossRef]
- Hackett, A.J.; Manning, J.S. Comments on feline syncytia-forming virus. J. Am. Vet. Med. Assoc. 1971, 158. Suppl 2: 948+. [Google Scholar]
- Helps, C.R.; Harbour, D.A. Comparison of the complete sequence of feline spumavirus with those of the primate spumaviruses reveals a shorter gag gene. J. Gen. Virol. 1997, 78, 2549–2564. [Google Scholar]
- Winkler, I.G.; Flugel, R.M.; Lochelt, M.; Flower, R.L. Detection and molecular characterisation of feline foamy virus serotypes in naturally infected cats. Virology 1998, 247, 144–151. [Google Scholar] [CrossRef]
- Zemba, M.; Alke, A.; Bodem, J.; Winkler, I.G.; Flower, R.L.; Pfrepper, K.; Delius, H.; Flugel, R.M.; Lochelt, M. Construction of infectious feline foamy virus genomes: Cat antisera do not cross-neutralize feline foamy virus chimera with serotype-specific env sequences. Virology 2000, 266, 150–156. [Google Scholar] [CrossRef]
- Löchelt, M. Personal communication. DKFZ: Heidelberg, Baden-Württemberg, Germany, 2013. [Google Scholar]
- Johnson, W.E.; Eizirik, E.; Pecon-Slattery, J.; Murphy, W.J.; Antunes, A.; Teeling, E.; O'Brien, S.J. The late miocene radiation of modern felidae: A genetic assessment. Science 2006, 311, 73–77. [Google Scholar] [CrossRef]
- O'Brien, S.J.; Johnson, W.E. Big cat genomics. Annu. Rev. Genom. Hum. Genet. 2005, 6, 407–429. [Google Scholar] [CrossRef]
- O'Brien, S.J.; Johnson, W.E. The evolution of cats. Genomic paw prints in the DNA of the world's wild cats have clarified the cat family tree and uncovered several remarkable migrations in their past. Sci. Am. 2007, 297, 68–75. [Google Scholar]
- Biomatters, Geneious, 6.05; Biomatters Ltd: New Zealand, 2013.
- Heads, M. Evolution and biogeography of primates: A new model based on molecular phylogenetics, vicariance and plate tectonics. Zoolog. Scripta 2010, 39, 107–127. [Google Scholar] [CrossRef]
- Bastone, P.; Truyen, U.; Lochelt, M. Potential of zoonotic transmission of non-primate foamy viruses to humans. J. Vet. Med. B 2003, 50, 417–423. [Google Scholar] [CrossRef]
- Calattini, S.; Betsem, E.B.; Froment, A.; Mauclere, P.; Tortevoye, P.; Schmitt, C.; Njouom, R.; Saib, A.; Gessain, A. Simian foamy virus transmission from apes to humans, rural cameroon. Emerg. Infect. Dis. 2007, 13, 1314–1320. [Google Scholar] [CrossRef]
- Murphy, H.W.; Miller, M.; Ramer, J.; Travis, D.; Barbiers, R.; Wolfe, N.D.; Switzer, W.M. Implications of simian retroviruses for captive primate population management and the occupational safety of primate handlers. J. Zoo Wildl. Med. 2006, 37, 219–233. [Google Scholar] [CrossRef]
- Locatelli, S.; Peeters, M. Cross-species transmission of simian retroviruses: How and why they could lead to the emergence of new diseases in the human population. Aids 2012, 26, 659–673. [Google Scholar] [CrossRef]
- Campbell, M.; Renshaw-Gegg, L.; Renne, R.; Luciw, P.A. Characterization of the internal promoter of simian foamy viruses. J. Virol. 1994, 68, 4811–4820. [Google Scholar]
- Lochelt, M.; Flugel, R.M.; Aboud, M. The human foamy virus internal promoter directs the expression of the functional bel 1 transactivator and bet protein early after infection. J. Virol. 1994, 68, 638–645. [Google Scholar]
- Lochelt, M. Foamy virus transactivation and gene expression. Curr. Top. Microbiol. Immunol. 2003, 277, 27–61. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. Meme suite: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lochelt, M.; Muranyi, W.; Flugel, R.M. Human foamy virus genome possesses an internal, bel-1-dependent and functional promoter. Proc. Natl. Acad. Sci. USA 1993, 90, 7317–7321. [Google Scholar] [CrossRef]
- Pfrepper, K.I.; Lochelt, M.; Rackwitz, H.R.; Schnolzer, M.; Heid, H.; Flugel, R.M. Molecular characterization of proteolytic processing of the gag proteins of human spumavirus. J. Virol. 1999, 73, 7907–7911. [Google Scholar]
- Enssle, J.; Fischer, N.; Moebes, A.; Mauer, B.; Smola, U.; Rethwilm, A. Carboxy-terminal cleavage of the human foamy virus gag precursor molecule is an essential step in the viral life cycle. J. Virol. 1997, 71, 7312–7317. [Google Scholar]
- Eastman, S.W.; Linial, M.L. Identification of a conserved residue of foamy virus gag required for intracellular capsid assembly. J. Virol. 2001, 75, 6857–6864. [Google Scholar] [CrossRef]
- Lindemann, D.; Pietschmann, T.; Picard-Maureau, M.; Berg, A.; Heinkelein, M.; Thurow, J.; Knaus, P.; Zentgraf, H.; Rethwilm, A. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 2001, 75, 5762–5771. [Google Scholar] [CrossRef]
- Geiselhart, V.; Bastone, P.; Kempf, T.; Schnolzer, M.; Lochelt, M. Furin-mediated cleavage of the feline foamy virus env leader protein. J. Virol. 2004, 78, 13573–13581. [Google Scholar] [CrossRef]
- Henikoff, S.; Henikoff, J.G. Amino acid substitution matrices from protein blocks. Proc. Natl. Acad. Sci. USA 1992, 89, 10915–10919. [Google Scholar] [CrossRef]
- Needleman, S.B.; Wunsch, C.D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 1970, 48, 443–453. [Google Scholar] [CrossRef]
- Thomas, G. Furin at the cutting edge: From protein traffic to embryogenesis and disease. Nature reviews. Molecular cell biology 2002, 3, 753–766. [Google Scholar] [CrossRef]
- Linial, M.L. Foamy viruses are unconventional retroviruses. J. Virol. 1999, 73, 1747–1755. [Google Scholar]
- Sa-Nguanmoo, P.; Rianthavorn, P.; Amornsawadwattana, S.; Poovorawan, Y. Hepatitis b virus infection in non-human primates. Acta Virol. 2009, 53, 73–82. [Google Scholar] [CrossRef]
- Rethwilm, A. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 2003, 277, 1–26. [Google Scholar] [CrossRef]
- Roy, J.; Rudolph, W.; Juretzek, T.; Gartner, K.; Bock, M.; Herchenroder, O.; Lindemann, D.; Heinkelein, M.; Rethwilm, A. Feline foamy virus genome and replication strategy. J. Virol. 2003, 77, 11324–11331. [Google Scholar] [CrossRef]
- Clarke, J.K.; Attridge, J.T. The morphology of simian foamy agents. J. Gen. Virol. 1968, 3, 185–190. [Google Scholar] [CrossRef]
- Clarke, J.K.; Attridge, J.T.; Gay, F.W. The morphogenesis of simian foamy agents. J. Gen. Virol. 1969, 4, 183–188. [Google Scholar] [CrossRef]
- Dermott, E.; Clarke, J.K.; Samuels, J. The morphogenesis and classification of bovine syncytial virus. J. Gen. Virol. 1971, 12, 105–119. [Google Scholar] [CrossRef]
- Riggs, J.L.; Oshiro, L.S.; Taylor, D.O.N.; Lennette, E.H. Syncytium-forming agent isolated from domestic cats. Nature 1969, 222, 1190–1191. [Google Scholar] [CrossRef]
- Flugel, R.M.; Pfrepper, K.I. Proteolytic processing of foamy virus gag and pol proteins. Curr. Top. Microbiol. Immunol. 2003, 277, 63–88. [Google Scholar] [CrossRef]
- Fischer, N.; Heinkelein, M.; Lindemann, D.; Enssle, J.; Baum, C.; Werder, E.; Zentgraf, H.; Muller, J.G.; Rethwilm, A. Foamy virus particle formation. J. Virol. 1998, 72, 1610–1615. [Google Scholar]
- Morozov, V.A.; Copeland, T.D.; Nagashima, K.; Gonda, M.A.; Oroszlan, S. Protein composition and morphology of human foamy virus intracellular cores and extracellular particles. Virology 1997, 228, 307–317. [Google Scholar] [CrossRef]
- Konvalinka, J.; Lochelt, M.; Zentgraf, H.; Flugel, R.M.; Krausslich, H.G. Active foamy virus proteinase is essential for virus infectivity but not for formation of a pol polyprotein. J. Virol. 1995, 69, 7264–7268. [Google Scholar]
- Zemba, M.; Wilk, T.; Rutten, T.; Wagner, A.; Flugel, R.M.; Lochelt, M. The carboxy-terminal p3gag domain of the human foamy virus gag precursor is required for efficient virus infectivity. Virology 1998, 247, 7–13. [Google Scholar] [CrossRef]
- Pincetic, A.; Leis, J. The mechanism of budding of retroviruses from cell membranes. Adv. Virol. 2009, 2009, 6239691–6239699. [Google Scholar]
- Life, R.B.; Lee, E.G.; Eastman, S.W.; Linial, M.L. Mutations in the amino terminus of foamy virus gag disrupt morphology and infectivity but do not target assembly. J. Virol. 2008, 82, 6109–6119. [Google Scholar] [CrossRef]
- Duda, A.; Stange, A.; Luftenegger, D.; Stanke, N.; Westphal, D.; Pietschmann, T.; Eastman, S.W.; Linial, M.L.; Rethwilm, A.; Lindemann, D. Prototype foamy virus envelope glycoprotein leader peptide processing is mediated by a furin-like cellular protease, but cleavage is not essential for viral infectivity. J. Virol. 2004, 78, 13865–13870. [Google Scholar] [CrossRef]
- Lindemann, D.; Goepfert, P.A. The foamy virus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 2003, 277, 111–129. [Google Scholar] [CrossRef]
- Stanke, N.; Stange, A.; Luftenegger, D.; Zentgraf, H.; Lindemann, D. Ubiquitination of the prototype foamy virus envelope glycoprotein leader peptide regulates subviral particle release. J. Virol. 2005, 79, 15074–15083. [Google Scholar] [CrossRef]
- Stange, A.; Luftenegger, D.; Reh, J.; Weissenhorn, W.; Lindemann, D. Subviral particle release determinants of prototype foamy virus. J. Virol. 2008, 82, 9858–9869. [Google Scholar] [CrossRef]
- Löchelt, M.; Bleiholder, A.; Wirthschaft, P. Personal communication. DKFZ: Heidelberg, Baden-Württemberg, Germany, 2013. [Google Scholar]
- Goepfert, P.A.; Shaw, K.L.; Ritter, G.D., Jr.; Mulligan, M.J. A sorting motif localizes the foamy virus glycoprotein to the endoplasmic reticulum. J. Virol. 1997, 71, 778–784. [Google Scholar]
- Goepfert, P.A.; Shaw, K.; Wang, G.; Bansal, A.; Edwards, B.H.; Mulligan, M.J. An endoplasmic reticulum retrieval signal partitions human foamy virus maturation to intracytoplasmic membranes. J. Virol. 1999, 73, 7210–7217. [Google Scholar]
- Jackson, M.R.; Nilsson, T.; Peterson, P.A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990, 9, 3153–3162. [Google Scholar]
- Goepfert, P.A.; Wang, G.; Mulligan, M.J. Identification of an er retrieval signal in a retroviral glycoprotein. Cell 1995, 82, 543–544. [Google Scholar] [CrossRef]
- Kong, X.H.; Yu, H.; Xuan, C.H.; Wang, J.Z.; Chen, Q.M.; Geng, Y.Q. The requirements and mechanism for capsid assembly and budding of bovine foamy virus. Arch. Virol. 2005, 150, 1677–1684. [Google Scholar] [CrossRef]
- Lecellier, C.H.; Neves, M.; Giron, M.L.; Tobaly-Tapiero, J.; Saib, A. Further characterization of equine foamy virus reveals unusual features among the foamy viruses. J. Virol. 2002, 76, 7220–7227. [Google Scholar] [CrossRef]
- Alke, A.; Schwantes, A.; Kido, K.; Flotenmeyer, M.; Flugel, R.M.; Lochelt, M. The bet gene of feline foamy virus is required for virus replication. Virology 2001, 287, 310–320. [Google Scholar] [CrossRef]
- Löchelt, M. DKFZ: Heidelberg, Baden-Württemberg, Germany, Unpublished work; 2010–2013.
- He, F.; Blair, W.S.; Fukushima, J.; Cullen, B.R. The human foamy virus bel-1 transcription factor is a sequence-specific DNA binding protein. J. Virol. 1996, 70, 3902–3908. [Google Scholar]
- Tan, J.; Hao, P.; Jia, R.; Yang, W.; Liu, R.; Wang, J.; Xi, Z.; Geng, Y.; Qiao, W. Identification and functional characterization of btas transactivator as a DNA-binding protein. Virology 2010, 405, 408–413. [Google Scholar] [CrossRef]
- Lochelt, M.; Aboud, M.; Flugel, R.M. Increase in the basal transcriptional activity of the human foamy virus internal promoter by the homologous long terminal repeat promoter in cis. Nucleic Acids Res. 1993, 21, 4226–4230. [Google Scholar] [CrossRef]
- Meiering, C.D.; Rubio, C.; May, C.; Linial, M.L. Cell-type-specific regulation of the two foamy virus promoters. J. Virol. 2001, 75, 6547–6557. [Google Scholar] [CrossRef]
- Erlwein, O.; Rethwilm, A. Bel-1 transactivator responsive sequences in the long terminal repeat of human foamy virus. Virology 1993, 196, 256–268. [Google Scholar] [CrossRef]
- Mergia, A.; Pratt-Lowe, E.; Shaw, K.E.; Renshaw-Gegg, L.W.; Luciw, P.A. Cis-acting regulatory regions in the long terminal repeat of simian foamy virus type 1. J. Virol. 1992, 66, 251–257. [Google Scholar]
- Renne, R.; Mergia, A.; Renshaw-Gegg, L.W.; Neumann-Haefelin, D.; Luciw, P.A. Regulatory elements in the long terminal repeat (ltr) of simian foamy virus type 3 (sfv-3). Virology 1993, 192, 365–369. [Google Scholar] [CrossRef]
- Omoto, S.; Brisibe, E.A.; Okuyama, H.; Fujii, Y.R. Feline foamy virus tas protein is a DNA-binding transactivator. J. Gen. Virol. 2004, 85, 2931–2935. [Google Scholar] [CrossRef]
- Liu, J.; Wang, S.; Zhang, L.; Qiao, W.; Chen, Q.; Geng, Y. Functional analysis of bovine foamy virus accessory protein and borf -1 responsive elements in long terminal repeat. Chin. J. Virol. 2000, 16, 141–149. [Google Scholar]
- Kang, Y.; Blair, W.S.; Cullen, B.R. Identification and functional characterization of a high-affinity bel-1 DNA binding site located in the human foamy virus internal promoter. J. Virol. 1998, 72, 504–511. [Google Scholar]
- Zou, J.X.; Luciw, P.A. The transcriptional transactivator of simian foamy virus 1 binds to a DNA target element in the viral internal promoter. Proc. Natl. Acad. Sci. USA 1996, 93, 326–330. [Google Scholar] [CrossRef]
- Bodem, J.; Kang, Y.; Flugel, R.M. Comparative functional characterization of the feline foamy virus transactivator reveals its species specificity. Virology 2004, 318, 32–36. [Google Scholar] [CrossRef]
- Flugel, R.M.; Rethwilm, A.; Maurer, B.; Darai, G. Nucleotide sequence analysis of the env gene and its flanking regions of the human spumaretrovirus reveals two novel genes. EMBO J. 1987, 6, 2077–2084. [Google Scholar]
- Mergia, A.; Shaw, K.E.; Pratt-Lowe, E.; Barry, P.A.; Luciw, P.A. Identification of the simian foamy virus transcriptional transactivator gene (taf). J. Virol. 1991, 65, 2903–2909. [Google Scholar]
- He, F.; Sun, J.D.; Garrett, E.D.; Cullen, B.R. Functional organization of the bel-1 trans activator of human foamy virus. J. Virol. 1993, 67, 1896–1904. [Google Scholar]
- Mergia, A.; Renshaw-Gegg, L.W.; Stout, M.W.; Renne, R.; Herchenroeder, O. Functional domains of the simian foamy virus type 1 transcriptional transactivator (taf). J. Virol. 1993, 67, 4598–4604. [Google Scholar]
- Blair, W.S.; Bogerd, H.; Cullen, B.R. Genetic analysis indicates that the human foamy virus bel-1 protein contains a transcription activation domain of the acidic class. J. Virol. 1994, 68, 3803–3808. [Google Scholar]
- Keller, A.; Partin, K.M.; Lochelt, M.; Bannert, H.; Flugel, R.M.; Cullen, B.R. Characterization of the transcriptional trans activator of human foamy retrovirus. J. Virol. 1991, 65, 2589–2594. [Google Scholar]
- Lee, C.W.; Chang, J.; Lee, K.J.; Sung, Y.C. The bel1 protein of human foamy virus contains one positive and two negative control regions which regulate a distinct activation domain of 30 amino acids. J. Virol. 1994, 68, 2708–2719. [Google Scholar]
- Tan, J.; Qiao, W.; Xu, F.; Han, H.; Chen, Q.; Geng, Y. Dimerization of btas is required for the transactivational activity of bovine foamy virus. Virology 2008, 376, 236–241. [Google Scholar] [CrossRef]
- Chang, J.; Lee, K.J.; Jang, K.L.; Lee, E.K.; Baek, G.H.; Sung, Y.C. Human foamy virus bel1 transactivator contains a bipartite nuclear localization determinant which is sensitive to protein context and triple multimerization domains. J. Virol. 1995, 69, 801–808. [Google Scholar]
- Qiao, W.; Tan, J. College of Life Sciences, Nankai University: Tianjin, China, Unpublished work; 2012.
- Bannert, H.; Muranyi, W.; Ogryzko, V.V.; Nakatani, Y.; Flugel, R.M. Coactivators p300 and pcaf physically and functionally interact with the foamy viral trans-activator. BMC Mol. Biol. 2004, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Bodem, J.; Krausslich, H.G.; Rethwilm, A. Acetylation of the foamy virus transactivator tas by pcaf augments promoter-binding affinity and virus transcription. J. Gen. Virol. 2007, 88, 259–263. [Google Scholar] [CrossRef]
- Chang, R.; Tan, J.; Xu, F.; Han, H.; Geng, Y.; Li, Y.; Qiao, W. Lysine acetylation sites in bovine foamy virus transactivator btas are important for its DNA binding activity. Virology 2011, 418, 21–26. [Google Scholar] [CrossRef]
- Wagner, A.; Doerks, A.; Aboud, M.; Alonso, A.; Tokino, T.; Flugel, R.M.; Lochelt, M. Induction of cellular genes is mediated by the bel1 transactivator in foamy virus-infected human cells. J. Virol. 2000, 74, 4441–4447. [Google Scholar] [CrossRef]
- Zheng, Y.H.; Jeang, K.T.; Tokunaga, K. Host restriction factors in retroviral infection: Promises in virus-host interaction. Retrovirology 2012, 9, 112. [Google Scholar] [CrossRef]
- Duggal, N.K.; Emerman, M. Evolutionary conflicts between viruses and restriction factors shape immunity. Nat. Rev. Immunol. 2012, 12, 687–695. [Google Scholar] [CrossRef]
- Zielonka, J.; Munk, C. Cellular restriction factors of feline immunodeficiency virus. Viruses 2011, 3, 1986–2005. [Google Scholar] [CrossRef]
- Harris, R.S.; Hultquist, J.F.; Evans, D.T. The restriction factors of human immunodeficiency virus. J. Biol. Chem. 2012, 287, 40875–40883. [Google Scholar]
- Zhang, C.; de Silva, S.; Wang, J.H.; Wu, L. Co-evolution of primate samhd1 and lentivirus vpx leads to the loss of the vpx gene in hiv-1 ancestor. PLoS One 2012, 7, e37477. [Google Scholar] [CrossRef]
- Chen, Z.; Zhang, L.; Ying, S. Samhd1: A novel antiviral factor in intrinsic immunity. Future Microbiol. 2012, 7, 1117–1126. [Google Scholar] [CrossRef]
- Holmes, R.K.; Malim, M.H.; Bishop, K.N. Apobec-mediated viral restriction: Not simply editing? Trends Biochem. Sci. 2007, 32, 118–128. [Google Scholar] [CrossRef]
- Zhang, J.; Ge, W.; Zhan, P.; De Clercq, E.; Liu, X. Retroviral restriction factors trim5alpha: Therapeutic strategy to inhibit hiv-1 replication. Curr. Med. Chem. 2011, 18, 2649–2654. [Google Scholar] [CrossRef]
- Kuhl, B.D.; Cheng, V.; Wainberg, M.A.; Liang, C. Tetherin and its viral antagonists. J. Neuroimmune Pharmacol. 2011, 6, 188–201. [Google Scholar] [CrossRef]
- Lilly, F. Susceptibility to two strains of friend leukemia virus in mice. Science 1967, 155, 461–462. [Google Scholar] [CrossRef]
- Laguette, N.; Benkirane, M. How samhd1 changes our view of viral restriction. Trends Immunol. 2012, 33, 26–33. [Google Scholar] [CrossRef]
- Sheehy, A.M.; Gaddis, N.C.; Choi, J.D.; Malim, M.H. Isolation of a human gene that inhibits hiv-1 infection and is suppressed by the viral vif protein. Nature 2002, 418, 646–650. [Google Scholar] [CrossRef]
- Yap, M.W.; Lindemann, D.; Stanke, N.; Reh, J.; Westphal, D.; Hanenberg, H.; Ohkura, S.; Stoye, J.P. Restriction of foamy viruses by primate trim5alpha. J. Virol. 2008, 82, 5429–5439. [Google Scholar] [CrossRef]
- Xu, F.; Tan, J.; Liu, R.; Xu, D.; Li, Y.; Geng, Y.; Liang, C.; Qiao, W. Tetherin inhibits prototypic foamy virus release. Virol. J. 2011, 8, 198. [Google Scholar] [CrossRef]
- Jouvenet, N.; Neil, S.J.; Zhadina, M.; Zang, T.; Kratovac, Z.; Lee, Y.; McNatt, M.; Hatziioannou, T.; Bieniasz, P.D. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 2009, 83, 1837–1844. [Google Scholar] [CrossRef]
- Lochelt, M.; Romen, F.; Bastone, P.; Muckenfuss, H.; Kirchner, N.; Kim, Y.B.; Truyen, U.; Rosler, U.; Battenberg, M.; Saib, A.; et al. The antiretroviral activity of apobec3 is inhibited by the foamy virus accessory bet protein. Proc. Natl. Acad. Sci. USA 2005, 102, 7982–7987. [Google Scholar] [CrossRef]
- Russell, R.A.; Wiegand, H.L.; Moore, M.D.; Schafer, A.; McClure, M.O.; Cullen, B.R. Foamy virus bet proteins function as novel inhibitors of the apobec3 family of innate antiretroviral defense factors. J. Virol. 2005, 79, 8724–8731. [Google Scholar] [CrossRef]
- Munk, C.; Beck, T.; Zielonka, J.; Hotz-Wagenblatt, A.; Chareza, S.; Battenberg, M.; Thielebein, J.; Cichutek, K.; Bravo, I.G.; O'Brien, S.J.; et al. Functions, structure, and read-through alternative splicing of feline apobec3 genes. Genome Biol. 2008, 9, R48. [Google Scholar] [CrossRef]
- Delebecque, F.; Suspene, R.; Calattini, S.; Casartelli, N.; Saib, A.; Froment, A.; Wain-Hobson, S.; Gessain, A.; Vartanian, J.P.; Schwartz, O. Restriction of foamy viruses by apobec cytidine deaminases. J. Virol. 2006, 80, 605–614. [Google Scholar] [CrossRef]
- Bransteitter, R.; Prochnow, C.; Chen, X.S. The current structural and functional understanding of apobec deaminases. Cell. Mol. Life Sci. 2009, 66, 3137–3147. [Google Scholar] [CrossRef]
- Smith, H.C.; Bennett, R.P.; Kizilyer, A.; McDougall, W.M.; Prohaska, K.M. Functions and regulation of the apobec family of proteins. Semin. Cell Dev. Biol. 2012, 23, 258–268. [Google Scholar] [CrossRef]
- Vartanian, J.P.; Guetard, D.; Henry, M.; Wain-Hobson, S. Evidence for editing of human papillomavirus DNA by apobec3 in benign and precancerous lesions. Science 2008, 320, 230–233. [Google Scholar] [CrossRef]
- Turelli, P.; Mangeat, B.; Jost, S.; Vianin, S.; Trono, D. Inhibition of hepatitis b virus replication by apobec3g. Science 2004, 303, 1829. [Google Scholar] [CrossRef]
- Chareza, S.; Slavkovic Lukic, D.; Liu, Y.; Rathe, A.M.; Munk, C.; Zabogli, E.; Pistello, M.; Lochelt, M. Molecular and functional interactions of cat apobec3 and feline foamy and immunodeficiency virus proteins: Different ways to counteract host-encoded restriction. Virology 2012, 424, 138–146. [Google Scholar] [CrossRef]
- Schafer, A.; Bogerd, H.P.; Cullen, B.R. Specific packaging of apobec3g into hiv-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 2004, 328, 163–168. [Google Scholar] [CrossRef]
- Perkovic, M.; Schmidt, S.; Marino, D.; Russell, R.A.; Stauch, B.; Hofmann, H.; Kopietz, F.; Kloke, B.P.; Zielonka, J.; Strover, H.; et al. Species-specific inhibition of apobec3c by the prototype foamy virus protein bet. J. Biol. Chem. 2009, 284, 5819–5826. [Google Scholar]
- Wissing, S.; Galloway, N.L.; Greene, W.C. Hiv-1 vif versus the apobec3 cytidine deaminases: An intracellular duel between pathogen and host restriction factors. Mol. Aspect. Med. 2010, 31, 383–397. [Google Scholar] [CrossRef]
- Mariani, R.; Chen, D.; Schrofelbauer, B.; Navarro, F.; Konig, R.; Bollman, B.; Munk, C.; Nymark-McMahon, H.; Landau, N.R. Species-specific exclusion of apobec3g from hiv-1 virions by vif. Cell 2003, 114, 21–31. [Google Scholar] [CrossRef]
- Romen, F.; Pawlita, M.; Sehr, P.; Bachmann, S.; Schroder, J.; Lutz, H.; Lochelt, M. Antibodies against gag are diagnostic markers for feline foamy virus infections while env and bet reactivity is undetectable in a substantial fraction of infected cats. Virology 2006, 345, 502–508. [Google Scholar] [CrossRef]
- Munk, C.; Willemsen, A.; Bravo, I.G. An ancient history of gene duplications, fusions and losses in the evolution of apobec3 mutators in mammals. BMC Evol. Biol. 2012, 12, 71. [Google Scholar]
- Hill, C.L.; Bieniasz, P.D.; McClure, M.O. Properties of human foamy virus relevant to its development as a vector for gene therapy. J. Gen. Virol. 1999, 80, 2003–2009. [Google Scholar]
- Mergia, A.; Leung, N.J.; Blackwell, J. Cell tropism of the simian foamy virus type 1 (sfv-1). J. Med. Primatol. 1996, 25, 2–7. [Google Scholar] [CrossRef]
- Phung, H.T.; Tohya, Y.; Shimojima, M.; Kato, K.; Miyazawa, T.; Akashi, H. Establishment of a gfp-based indicator cell line to quantitate feline foamy virus. J. Virol. Meth. 2003, 109, 125–131. [Google Scholar] [CrossRef]
- Mikovits, J.A.; Hoffman, P.M.; Rethwilm, A.; Ruscetti, F.W. In vitro infection of primary and retrovirus-infected human leukocytes by human foamy virus. J. Virol. 1996, 70, 2774–2780. [Google Scholar]
- Malmquist, W.A.; Van der Maaten, M.J.; Boothe, A.D. Isolation, immunodiffusion, immunofluorescence, and electron microscopy of a syncytial virus of lymphosarcomatous and apparently normal cattle. Canc. Res. 1969, 29, 188–200. [Google Scholar]
- Hatama, S.; Otake, K.; Ohta, M.; Kobayashi, M.; Imakawa, K.; Ikemoto, A.; Okuyama, H.; Mochizuki, M.; Miyazawa, T.; Tohya, Y.; et al. Reactivation of feline foamy virus from a chronically infected feline renal cell line by trichostatin a. Virology 2001, 283, 315–323. [Google Scholar] [CrossRef]
- Liebermann, H.; Riebe, R. Isolation of bovine syncytial virus in East Germany. Arch. Exp. Veterinarmed. 1981, 35, 917–919. [Google Scholar]
- Ma, Z.; Hao, P.; Yao, X.; Liu, C.; Tan, J.; Liu, L.; Yang, R.; Geng, Y.; Chen, Q.; Qiao, W. Establishment of an indicator cell line to quantify bovine foamy virus infection. J. Basic Microbiol. 2008, 48, 278–283. [Google Scholar] [CrossRef]
- Yu, S.F.; Linial, M.L. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J. Virol. 1993, 67, 6618–6624. [Google Scholar]
- von Laer, D.; Neumann-Haefelin, D.; Heeney, J.L.; Schweizer, M. Lymphocytes are the major reservoir for foamy viruses in peripheral blood. Virology 1996, 221, 240–244. [Google Scholar] [CrossRef]
- Callahan, M.E.; Switzer, W.M.; Matthews, A.L.; Roberts, B.D.; Heneine, W.; Folks, T.M.; Sandstrom, P.A. Persistent zoonotic infection of a human with simian foamy virus in the absence of an intact orf-2 accessory gene. J. Virol. 1999, 73, 9619–9624. [Google Scholar]
- German, A.C.; Harbour, D.A.; Helps, C.R.; Gruffydd-Jones, T.J. Is feline foamy virus really apathogenic? Vet. Immunol. Immunopathol. 2008, 123, 114–118. [Google Scholar] [CrossRef]
- Materniak, M.; Hechler, T.; Lochelt, M.; Kuzmak, J. Similar patterns of infection with bovine foamy virus in experimentally inoculated calves and sheep. J. Virol. 2013, 87, 3516–3525. [Google Scholar] [CrossRef]
- Bouillant, A.M.; Ruckerbauer, G.M. Isolation of bovine syncytial virus from lymphocytes recovered from fluids used to flush uterus and oviducts of superovulated cattle. Can. J. Comp. Med. 1984, 48, 332–334. [Google Scholar]
- Scott, F.W.; Shively, J.N.; Gaskin, J.; Gillespie, J.H. Bovine syncytial virus isolations. Arch. Gesamte Virusforsch. 1973, 43, 43–52. [Google Scholar] [CrossRef]
- Van der Maaten, M.J.; Hubbert, W.T.; Boothe, A.D.; Bryner, J.H.; Estes, P.C. Isolations of bovine syncytial virus from maternal and fetal blood. Am. J. Vet. Res. 1973, 34, 341–343. [Google Scholar]
- Greig, A.S. A syncytium regression test to detect antibodies to bovine syncytia virus. Can. J. Comp. Med. 1979, 43, 112–114. [Google Scholar]
- Luther, P.D.; Nuttall, P.A.; Gibbons, R.A. Isolation of viruses from cultures of bovine endometrial cells. J. Infect. Dis. 1978, 138, 660–663. [Google Scholar] [CrossRef]
- Granzow, H.; Liebermann, H.; Riebe, R.; Solisch, P. Morphology and morphogenesis of bovine syncytial virus--electron microscopy of ultramicrotomic sections. Arch. Exp. Veterinarmed. 1981, 35, 791–800. [Google Scholar]
- Falcone, V.; Leupold, J.; Clotten, J.; Urbanyi, E.; Herchenroder, O.; Spatz, W.; Volk, B.; Bohm, N.; Toniolo, A.; Neumann-Haefelin, D.; et al. Sites of simian foamy virus persistence in naturally infected african green monkeys: Latent provirus is ubiquitous, whereas viral replication is restricted to the oral mucosa. Virology 1999, 257, 7–14. [Google Scholar] [CrossRef]
- Murray, S.M.; Picker, L.J.; Axthelm, M.K.; Linial, M.L. Expanded tissue targets for foamy virus replication with simian immunodeficiency virus-induced immunosuppression. J. Virol. 2006, 80, 663–670. [Google Scholar] [CrossRef]
- Materniak, M.H.T.; Löchelt, M.; Kuzmak, J. Seroreactivity of humans and ruminants to bovine foamy virus—Evidence for zoonotic transmission and existence of new reservoirs for foamy viruses. In Proceedings of the 8th International Foamy Virus Conference, Argos, Greece, 7–8 May 2010.
- Nakamura, K.; Miyazawa, T.; Ikeda, Y.; Sato, E.; Nishimura, Y.; Nguyen, N.T.; Takahashi, E.; Mochizuki, M.; Mikami, T. Contrastive prevalence of feline retrovirus infections between northern and southern vietnam. J. Vet. Med. Sci. 2000, 62, 921–923. [Google Scholar] [CrossRef]
- Bandecchi, P.; Matteucci, D.; Baldinotti, F.; Guidi, G.; Abramo, F.; Tozzini, F.; Bendinelli, M. Prevalence of feline immunodeficiency virus and other retroviral infections in sick cats in italy. Vet. Immunol. Immunopathol. 1992, 31, 337–345. [Google Scholar] [CrossRef]
- Daniels, M.J.; Golder, M.C.; Jarrett, O.; MacDonald, D.W. Feline viruses in wildcats from scotland. J. Wildl. Dis. 1999, 35, 121–124. [Google Scholar]
- Glaus, T.; Hofmann-Lehmann, R.; Greene, C.; Glaus, B.; Wolfensberger, C.; Lutz, H. Seroprevalence of bartonella henselae infection and correlation with disease status in cats in switzerland. J. Clin. Microbiol. 1997, 35, 2883–2885. [Google Scholar]
- Winkler, I.G.; Lochelt, M.; Flower, R.L. Epidemiology of feline foamy virus and feline immunodeficiency virus infections in domestic and feral cats: A seroepidemiological study. J. Clin. Microbiol. 1999, 37, 2848–2851. [Google Scholar]
- Miyazawa, T.; Ikeda, Y.; Maeda, K.; Horimoto, T.; Tohya, Y.; Mochizuki, M.; Vu, D.; Vu, G.D.; Cu, D.X.; Ono, K.; et al. Seroepidemiological survey of feline retrovirus infections in domestic and leopard cats in northern vietnam in 1997. J. Vet. Med. Sci. 1998, 60, 1273–1275. [Google Scholar] [CrossRef]
- Bleiholder, A.; Muhle, M.; Hechler, T.; Bevins, S.; vandeWoude, S.; Denner, J.; Lochelt, M. Pattern of seroreactivity against feline foamy virus proteins in domestic cats from germany. Vet. Immunol. Immunopathol. 2011, 143, 292–300. [Google Scholar] [CrossRef]
- Löchelt, M.; Bleiholder, A. Personal communication. DKFZ: Heidelberg, Baden-Württemberg, Germany, 2013. [Google Scholar]
- Jacobs, R.M.; Pollari, F.L.; McNab, W.B.; Jefferson, B. A serological survey of bovine syncytial virus in ontario: Associations with bovine leukemia and immunodeficiency-like viruses, production records, and management practices. Can. J. Vet. Res. 1995, 59, 271–278. [Google Scholar]
- Johnson, R.H.; de la Rosa, J.; Abher, I.; Kertayadnya, I.G.; Entwistle, K.W.; Fordyce, G.; Holroyd, R.G. Epidemiological studies of bovine spumavirus. Vet. Microbiol. 1988, 16, 25–33. [Google Scholar] [CrossRef]
- Pamba, R.; Jeronimo, C.; Archambault, D. Detection of bovine retrospumavirus by the polymerase chain reaction. J. Virol. Meth. 1999, 78, 199–208. [Google Scholar] [CrossRef]
- Romen, F.; Backes, P.; Materniak, M.; Sting, R.; Vahlenkamp, T.W.; Riebe, R.; Pawlita, M.; Kuzmak, J.; Lochelt, M. Serological detection systems for identification of cows shedding bovine foamy virus via milk. Virology 2007, 364, 123–131. [Google Scholar] [CrossRef]
- Materniak, M. National Veterinary Research Institute: Pulawy, Poland, Unpublished work; 2007.
- Materniak, M.; Kuzmak, J. Occurrence of equine foamy virus infection in horses from poland. In Proceedings of the 9th International Foamy Virus Conference, Bethesda, MD, USA, 29–30 May 2012.
- Nasimuzzaman, M.; Persons, D.A. Cell membrane-associated heparan sulfate is a receptor for prototype foamy virus in human, monkey, and rodent cells. Mol. Ther. 2012, 20, 1158–1166. [Google Scholar] [CrossRef]
- Plochmann, K.; Horn, A.; Gschmack, E.; Armbruster, N.; Krieg, J.; Wiktorowicz, T.; Weber, C.; Stirnnagel, K.; Lindemann, D.; Rethwilm, A.; et al. Heparan sulfate is an attachment factor for foamy virus entry. J. Virol. 2012, 86, 10028–10035. [Google Scholar] [CrossRef]
- Calattini, S.; Wanert, F.; Thierry, B.; Schmitt, C.; Bassot, S.; Saib, A.; Herrenschmidt, N.; Gessain, A. Modes of transmission and genetic diversity of foamy viruses in a macaca tonkeana colony. Retrovirology 2006, 3, 23. [Google Scholar] [CrossRef]
- Alke, A.; Schwantes, A.; Zemba, M.; Flugel, R.M.; Lochelt, M. Characterization of the humoral immune response and virus replication in cats experimentally infected with feline foamy virus. Virology 2000, 275, 170–176. [Google Scholar] [CrossRef]
- Leendertz, F.H.; Zirkel, F.; Couacy-Hymann, E.; Ellerbrok, H.; Morozov, V.A.; Pauli, G.; Hedemann, C.; Formenty, P.; Jensen, S.A.; Boesch, C.; et al. Interspecies transmission of simian foamy virus in a natural predator-prey system. J. Virol. 2008, 82, 7741–7744. [Google Scholar] [CrossRef]
- Liu, W.; Worobey, M.; Li, Y.; Keele, B.F.; Bibollet-Ruche, F.; Guo, Y.; Goepfert, P.A.; Santiago, M.L.; Ndjango, J.B.; Neel, C.; et al. Molecular ecology and natural history of simian foamy virus infection in wild-living chimpanzees. PLoS Pathog. 2008, 4, e1000097. [Google Scholar] [CrossRef]
- Goffe, A.S.; Blasse, A.; Mundry, R.; Leendertz, F.H.; Calvignac-Spencer, S. Detection of retroviral super-infection from non-invasive samples. PLoS One 2012, 7, e36570. [Google Scholar]
- Galvin, T.A.; Ahmed, I.A.; Shahabuddin, M.; Bryan, T.; Khan, A.S. Identification of recombination in the envelope gene of simian foamy virus serotype 2 isolated from macaca cyclopis (sfvmcy-2). J. Virol. 2013, 87, 8792–8797. [Google Scholar] [CrossRef]
- Choudhary, A.; Galvin, T.A.; Williams, D.K.; Beren, J.; Bryant, M.A.; Khan, A.S. Influence of naturally occurring simian foamy viruses (sfvs) on siv disease progression in the rhesus macaque (macaca mulatta) model. Viruses 2013, 5, 1414–1430. [Google Scholar] [CrossRef]
- Amborski, G.F.; Storz, J.; Keney, D.; Lo, J.; McChesney, A.E. Isolation of a retrovirus from the american bison and its relation to bovine retroviruses. J. Wildl. Dis. 1987, 23, 7–11. [Google Scholar]
- Mochizuki, M.; Akuzawa, M.; Nagatomo, H. Serological survey of the iriomote cat (felis iriomotensis) in japan. J. Wildl. Dis. 1990, 26, 236–245. [Google Scholar]
- Mouinga-Ondeme, A.; Caron, M.; Nkoghe, D.; Telfer, P.; Marx, P.; Saib, A.; Leroy, E.; Gonzalez, J.P.; Gessain, A.; Kazanji, M. Cross-species transmission of simian foamy virus to humans in rural gabon, central africa. J. Virol. 2012, 86, 1255–1260. [Google Scholar] [CrossRef]
- Wolfe, N.D.; Switzer, W.M.; Carr, J.K.; Bhullar, V.B.; Shanmugam, V.; Tamoufe, U.; Prosser, A.T.; Torimiro, J.N.; Wright, A.; Mpoudi-Ngole, E.; et al. Naturally acquired simian retrovirus infections in central african hunters. Lancet 2004, 363, 932–937. [Google Scholar] [CrossRef]
- Betsem, E.; Rua, R.; Tortevoye, P.; Froment, A.; Gessain, A. Frequent and recent human acquisition of simian foamy viruses through apes' bites in central africa. PLoS Pathog. 2011, 7, e1002306. [Google Scholar] [CrossRef]
- Heneine, W.; Switzer, W.M.; Sandstrom, P.; Brown, J.; Vedapuri, S.; Schable, C.A.; Khan, A.S.; Lerche, N.W.; Schweizer, M.; Neumann-Haefelin, D.; et al. Identification of a human population infected with simian foamy viruses. Nat. Med. 1998, 4, 403–407. [Google Scholar] [CrossRef]
- Boneva, R.S.; Switzer, W.M.; Spira, T.J.; Bhullar, V.B.; Shanmugam, V.; Cong, M.E.; Lam, L.; Heneine, W.; Folks, T.M.; Chapman, L.E. Clinical and virological characterization of persistent human infection with simian foamy viruses. AIDS Res. Hum. Retrovir. 2007, 23, 1330–1337. [Google Scholar] [CrossRef]
- Butera, S.T.; Brown, J.; Callahan, M.E.; Owen, S.M.; Matthews, A.L.; Weigner, D.D.; Chapman, L.E.; Sandstrom, P.A. Survey of veterinary conference attendees for evidence of zoonotic infection by feline retroviruses. J. Am. Vet. Med. Assoc. 2000, 217, 1475–1479. [Google Scholar] [CrossRef]
- Winkler, I.G.; Lochelt, M.; Levesque, J.P.; Bodem, J.; Flugel, R.M.; Flower, R.L. A rapid streptavidin-capture elisa specific for the detection of antibodies to feline foamy virus. J. Immunol. Meth. 1997, 207, 69–77. [Google Scholar] [CrossRef]
- Hooks, J.J.; Burns, W.; Hayashi, K.; Geis, S.; Notkins, A.L. Viral spread in the presence of neutralizing antibody: Mechanisms of persistence in foamy virus infection. Infect. Immun. 1976, 14, 1172–1178. [Google Scholar]
- Hooks, J.J.; Gibbs, C.J., Jr. The foamy viruses. Bacteriol. Rev. 1975, 39, 169–185. [Google Scholar]
- Muhle, M.; Bleiholder, A.; Kolb, S.; Hubner, J.; Lochelt, M.; Denner, J. Immunological properties of the transmembrane envelope protein of the feline foamy virus and its use for serological screening. Virology 2011, 412, 333–340. [Google Scholar] [CrossRef]
- Hussain, A.I.; Shanmugam, V.; Bhullar, V.B.; Beer, B.E.; Vallet, D.; Gautier-Hion, A.; Wolfe, N.D.; Karesh, W.B.; Kilbourn, A.M.; Tooze, Z.; et al. Screening for simian foamy virus infection by using a combined antigen western blot assay: Evidence for a wide distribution among old world primates and identification of four new divergent viruses. Virology 2003, 309, 248–257. [Google Scholar] [CrossRef]
- Calattini, S.; Nerrienet, E.; Mauclere, P.; Georges-Courbot, M.C.; Saib, A.; Gessain, A. Natural simian foamy virus infection in wild-caught gorillas, mandrills and drills from cameroon and gabon. J. Gen. Virol. 2004, 85, 3313–3317. [Google Scholar] [CrossRef]
- Rua, R.; Betsem, E.; Calattini, S.; Saib, A.; Gessain, A. Genetic characterization of simian foamy viruses infecting humans. J. Virol. 2012, 86, 13350–13359. [Google Scholar] [CrossRef]
- Schweizer, M.; Neumann-Haefelin, D. Phylogenetic analysis of primate foamy viruses by comparison of pol sequences. Virology 1995, 207, 577–582. [Google Scholar] [CrossRef]
- Lew, A.E.; Bock, R.E.; Miles, J.; Cuttell, L.B.; Steer, P.; Nadin-Davis, S.A. Sensitive and specific detection of bovine immunodeficiency virus and bovine syncytial virus by 5' taq nuclease assays with fluorescent 3' minor groove binder-DNA probes. J. Virol. Meth. 2004, 116, 1–9. [Google Scholar] [CrossRef]
- Materniak, M.; Sieradzki, Z.; Kuzmak, J. Detection of bovine foamy virus in milk and saliva of bfv seropositive cattle. Bull. Vet. Inst. Pulawy. 2010, 54, 461–465. [Google Scholar]
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Kehl, T.; Tan, J.; Materniak, M. Non-Simian Foamy Viruses: Molecular Virology, Tropism and Prevalence and Zoonotic/Interspecies Transmission. Viruses 2013, 5, 2169-2209. https://doi.org/10.3390/v5092169
Kehl T, Tan J, Materniak M. Non-Simian Foamy Viruses: Molecular Virology, Tropism and Prevalence and Zoonotic/Interspecies Transmission. Viruses. 2013; 5(9):2169-2209. https://doi.org/10.3390/v5092169
Chicago/Turabian StyleKehl, Timo, Juan Tan, and Magdalena Materniak. 2013. "Non-Simian Foamy Viruses: Molecular Virology, Tropism and Prevalence and Zoonotic/Interspecies Transmission" Viruses 5, no. 9: 2169-2209. https://doi.org/10.3390/v5092169




