Perspective of Use of Antiviral Peptides against Influenza Virus
Abstract
:1. Introduction
2. Design and Characteristics of Antiviral Peptides
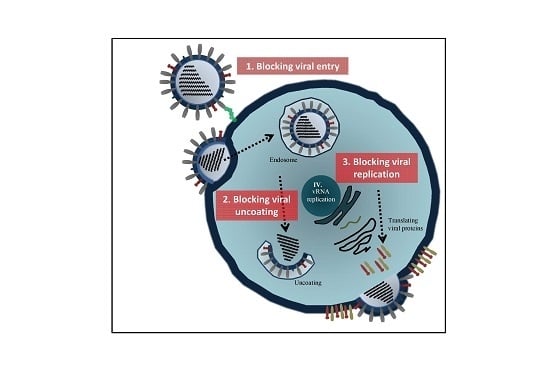
Influenza Virus Replication Cycle
3. Mode of Action of Various Antimicrobial Peptides with Antiviral Activity
3.1. The Peptides Inhibiting Virus Attachment and Virus-Cell Membrane Fusion


| The Peptides Inhibiting Virus Attachment and Virus-Cell Membrane Fusion | |||||||
| Peptide | Influenza Serotype | Sequence | Conformation | Net Charge * | Hydrophobic Residue * | IC50 | Reference |
| EB peptide | Broad spectrum | RRKKAAVALLPAVLLALLAP | linear | 4 | 70 | 3 to 20 µM | [37] |
| Derived EB peptide | Broad spectrum | RRKKLAVLLALLA | linear | 4 | 69 | 3.5 µM | [38] |
| P1 | H9N2 | NDFRSKT | linear | 1 | 14 | 48 µM | [39] |
| P1 cyclic | H9N3 | CNDFRSKTC | cyclic | 1 | 33 | 71 µM | [39] |
| FluPep 1 | H1N1 | WLVFFVIFYFFR | α-helix | 1 | 83 | 0.093 µM | [40] |
| FluPep 2 | H1N1 | WLVFFVIAYFAR | α-helix | 1 | 83 | 0.0009 µM | [40] |
| FluPep 3 | H1N1 | WLVFFVIFYFFRRRKK | α-helix | 5 | 62 | 0.00003 µM | [40] |
| FluPep 4 | H1N1 | RRKKWLVFFVIFYFFR | α-helix | 5 | 62 | 0.00004 µM | [40] |
| FluPep 7 | H1N1 | RRKKIFYFFR | α-helix | 5 | 40 | 0.15 µM | [40] |
| FluPep 8 | H1N1 | WLVFFVRRKK | α-helix | 4 | 60 | 0.63 µM | [40] |
| FluPep 9 | H1N1 | FFVIFYRRKK | α-helix | 4 | 50 | 1.48 µM | [40] |
| C18-s2 | H1N1, H3N2 | C17H35CO-ARLPRTMVHPKPAQP-NH2 | - | 3 | 33 | 11–15 µM | [41] |
| Pal L1 | H5N1 | C16-ARLPRTMVHPKPAQP | micelle | 3 | 33 | - | [42] |
| Pal M1 | H5N1 | C16-ARLPRTMV | micelle | 2 | 50 | - | [42] |
| Pal S1 | H5N1 | C16-ARLPR | micelle | 2 | 40 | - | [42] |
| Flufirvitide | Broad spectrum | - | - | - | - | - | [43] |
| PEP 19-2.5 | H7N7, H3N2, H1N1 | GCKKYRRFRWKFKGKFWFWG | α-helix | 8 | 40 | - | [44] |
| PEP 19-4 | H7N7, H3N2, H1N1 | GKKYRRFRWKFKGKWFWFG | α-helix | 8 | 36 | - | [44] |
| PEP 19-8D | H7N7, H3N2, H1N1 | GFWFKGKWRFKKYRGGRYKKFRWKGKFWFG | α-helix | 12 | 33 | - | [44] |
| PEP 19-CP | H7N7, H3N2, H1N1 | SSNKSTTGSGETTTA | α-helix | 0 | 6 | - | [44] |
| Defensins | H1N1, H3N2 | ACYCRIPACIAGERRYGTCIYQGRLWAFCC | β-sheet | 3 | 53 | - | [45] |
| The Peptides Disrupting Viral Envelope | |||||||
| Peptide | Influenza Serotype | Sequence | Conformation | Net Charge * | Hydrophobic Residue * | IC50 | Reference |
| LF C-lobe peptide 1 | H1H1, H3N2 | SKHSSLDCVLRP | α-helix | 1 | 33 | 4–6 pM | [46] |
| LF C-lobe peptide 2 | H1H1, H3N2 | AGDDQGLDKCVPNSKEK | α-helix | −1 | 23 | 4–7 pM | [46] |
| LF C-lobe peptide 3 | H1H1, H3N2 | NGESSADWAKN | α-helix | −1 | 27 | 22–225 pM | [46] |
| Mucroporin-M1 | H5N1, H1N1 | LFRLIKSLIKRLVSAFK | α-helix | 5 | 58 | 1.03 μM | [47] |
| LL-37 | H1N1, H3N2 | LLGDFFRKSKEKIGKEFKRIVQRIKDFLRNLVPRTES | α-helix | 6 | 35 | - | [48] |
| The Peptides Inhibiting Viral Replication | |||||||
| Peptide | Influenza Serotype | Sequence | Conformation | Net Charge* | Hydrophobic Residue* | IC50 | Reference |
| PB11-25 | Broad spectrum | MDVNPTLLFLKVPAQNAISTTFPYT | α-helix | 0 | 44 | - | [49] |
| PB21-37 | H1N1, H5N1 | MERIKELRDLMSWSRTREILTKTTVDHMAIIKKYTSG | α-helix | 3 | 35 | 375 nM | [50] |
| PB1731–757 | H5N1 | ESGRIKKEEFAEIMKICSTIEELGRQK | α-helix | 0 | 33 | - | [51] |
| PB11–25AT6Y | H1N1, H5N1 | MDVNPYLLFLKVPAQ | α-helix | 0 | 53 | 22–107 nM | [52] |
| Killer peptide | H7N1 | AKVTMTCSAS | α-helix | 1 | 50 | 2.6 µM | [53] |
| HNP-1 | H3N2 | CYCRIPACIAGERRYGTCIYQGRLWAFCC | β-sheet | 3 | 51 | - | [54] |
| Peptid 6 | H1N1, H3N2 | CATCEQIADSQHRSHRQMV | Zn-finger | 0 | 36 | 0.7 nM | [55,56] |
3.2. The Peptides Disrupting Viral Envelope

3.3. The Peptides Inhibiting Viral Replication
3.4. Other Possible Mechanisms of Influenza Virus Inhibition
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Wiesner, J.; Vilcinskas, A. Antimicrobial peptides the ancient arm of the human immune system. Virulence 2010, 1, 440–464. [Google Scholar] [CrossRef] [PubMed]
- Mandal, S.M.; Silva, O.N.; Franco, O.L. Recombinant probiotics with antimicrobial peptides: A dual strategy to improve immune response in immunocompromised patients. Drug Discov. Today 2014, 19, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Nizet, V.; Ohtake, T.; Lauth, X.; Trowbridge, J.; Rudisill, J.; Dorschner, R.A.; Pestonjamasp, V.; Piraino, J.; Huttner, K.; Gallo, R.L. Innate antimicrobial peptide protects the skin from invasive bacterial infection. Nature 2001, 414, 454–457. [Google Scholar] [CrossRef] [PubMed]
- Tew, G.N.; Clements, D.; Tang, H.Z.; Arnt, L.; Scott, R.W. Antimicrobial activity of an abiotic host defense peptide mimic. Biochim. Biophys. Acta 2006, 1758, 1387–1392. [Google Scholar] [CrossRef]
- Silva, R.R.; Avelino, K.; Ribeiro, K.L.; Franco, O.L.; Oliveira, M.D.L.; Andrade, C.A.S. Optical and dielectric sensors based on an microbial peptides for microorganism diagnosis. Front. Microbiol. 2014, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Hale, J.D.; Hancock, R.E. Alternative mechanisms of action of cationic antimicrobial peptides on bacteria. Expert Rev. Anti Infect. Ther. 2007, 5, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Hadley, E.B.; Hancock, R.E.W. Strategies for the discovery and advancement of novel cationic antimicrobial peptides. Curr. Top. Med. Chem. 2010, 10, 1872–1881. [Google Scholar] [CrossRef] [PubMed]
- Fosgerau, K.; Hoffmann, T. Peptide therapeutics: Current status and future directions. Drug Discov. Today 2015, 20, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Sim, I.S. Potential of new drugs in the prophylaxis and therapy of influenza A virus-infections. Curr. Opin. Infect. Dis. 1989, 2, 411–414. [Google Scholar] [CrossRef]
- Zumla, A.; Memish, Z.A.; Maeurer, M.; Bates, M.; Mwaba, P.; Al-Tawfiq, J.A.; Denning, D.W.; Hayden, F.G.; Hui, D.S. Emerging novel and antimicrobial-resistant respiratory tract infections: New drug development and therapeutic options. Lancet Infect. Dis. 2014, 14, 1136–1149. [Google Scholar] [CrossRef]
- Berkhout, B.; Sanders, R.W. Molecular strategies to design an escape-proof antiviral therapy. Antivir. Res. 2011, 92, 7–14. [Google Scholar] [PubMed]
- Wilson, J.C.; von Itzstein, M. Recent strategies in the search for new anti-influenza therapies. Curr. Drug Targets 2003, 4, 389–408. [Google Scholar] [CrossRef] [PubMed]
- Air, G.M.; Brouillette, W.J. Influenza virus antiviral targets. Antiviral Res. 2009, 187–207. [Google Scholar]
- Krol, E.; Rychowska, M.; Szewczyk, B. Antivirals—Current trends in fighting influenza. Acta Biochim. Pol. 2014, 61, 495–504. [Google Scholar] [PubMed]
- Jitendra, P.K.; Bansal, S.; Banik, A. Noninvasive routes of proteins and peptides drug delivery. Indian J. Pharm. Sci. 2011, 73, 367–375. [Google Scholar] [PubMed]
- Vlieghe, P.; Lisowski, V.; Martinez, J.; Khrestchatisky, M. Synthetic therapeutic peptides: Science and market. Drug Discov. Today 2010, 15, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Liderot, K.; Ahl, M.; Ozenci, V. Secondary bacterial infections in patients with seasonal influenza A and pandemic H1N1. Biomed. Res. Int. 2013, 2013, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, V.; Feio, M.J.; Bastos, M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog. Lipid Res. 2012, 51, 149–177. [Google Scholar] [CrossRef] [PubMed]
- Giangaspero, A.; Sandri, L.; Tossi, A. Amphipathic alpha helical antimicrobial peptides—A systematic study of the effects of structural and physical properties on biological activity. Eur. J. Biochem. 2001, 268, 5589–5600. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.M.; Edwards, M.A.; Li, J.; Yip, C.M.; Deber, C.M. Roles of hydrophobicity and charge distribution of cationic antimicrobial peptides in peptide-membrane interactions. J. Biol. Chem. 2012, 287, 7738–7745. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Walker, E.; Egerer, L.; Bunnell, B.A.; Mondal, D.; von Laer, D.; Braun, S.E. The therapeutic potential of secreted antiviral entry inhibitor (SAVE) peptides expressed by transduced MSCs to block HIV infection. Mol. Ther. 2014, 22, S183. [Google Scholar]
- Pessi, A.; Ingallinella, P.; Bianchi, E.; Wang, Y.J.; Hrin, R.; Veneziano, M.; Bonelli, F.; Ketas, T.; Moore, J.; Miller, M. Dramatic increase of antiviral potency of an HIV peptide fusion inhibitor by targeting to lipid rafts via addition of a cholesterol group. Biopolymers 2009, 92, 302. [Google Scholar]
- Abe, K.; Nozaki, A.; Tamura, K.; Ikeda, M.; Naka, K.; Dansako, H.; Hoshino, H.; Tanaka, K.; Kato, N. Tandem repeats of lactoferrin-derived anti-hepatitis C virus peptide enhance antiviral activity in cultured human hepatocytes. Microbiol. Immunol. 2007, 51, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Jenssen, H. Anti herpes simplex virus activity of lactoferrin/lactoferricin—An example of antiviral activity of antimicrobial protein/peptide. Cell. Mol. Life Sci. 2005, 62, 3002–3013. [Google Scholar] [CrossRef] [PubMed]
- Jaishankar, D.; Yakoub, A.M.; Bogdanov, A.; Valyi-Nagy, T.; Shukla, D. Characterization of a proteolytically stable d-peptide that suppresses herpes simplex virus 1 infection: Implications for the development of entry-based antiviral therapy. J. Virol. 2015, 89, 1932–1938. [Google Scholar] [CrossRef] [PubMed]
- Albericio, F.; Kruger, H.G. Therapeutic peptides foreword. Future Med. Chem. 2012, 4, 1527–1531. [Google Scholar] [CrossRef] [PubMed]
- She, R.; Bao, H.; Zhang, Y.; Luo, D. Effect of rabbit sacculus rotundus antimicrobial peptides on serum antibody titers after vaccination with newcastle disease virus vaccine and avian influenza virus vaccine in chickens. Poult. Sci. 2008, 87, 81–86. [Google Scholar]
- Pizzorno, A.; Abed, Y.; Boivin, G. Influenza drug resistance. Semin. Respir. Crit. Care Med. 2011, 32, 409–422. [Google Scholar] [CrossRef] [PubMed]
- Mikulasova, A.; Vareckova, E.; Fodor, E. Transcription and replication of the influenza A virus genome. Acta Virol. 2000, 44, 273–282. [Google Scholar] [PubMed]
- Das, K.; Aramini, J.M.; Ma, L.C.; Krug, R.M.; Arnold, E. Structures of influenza A proteins and insights into antiviral drug targets. Nat. Struct. Mol. Biol. 2010, 17, 530–538. [Google Scholar] [CrossRef] [PubMed]
- Brunotte, L.; Flies, J.; Bolte, H.; Reuther, P.; Vreede, F.; Schwemmle, M. The nuclear export protein of H5N1 influenza A viruses recruits matrix 1 (M1) protein to the viral ribonucleoprotein to mediate nuclear export. J. Biol. Chem. 2014, 289, 20067–20077. [Google Scholar] [CrossRef] [PubMed]
- Stubbs, T.M.; te Velthuis, A.J.W. The RNA-dependent RNA polymerase of the influenza A virus. Future Virol. 2014, 9, 863–876. [Google Scholar] [CrossRef] [PubMed]
- Leser, G.P.; Lamb, R.A. Influenza virus assembly and budding in raft-derived microdomains: A quantitative analysis of the surface distribution of HA, NA and M2 proteins. Virology 2005, 342, 215–227. [Google Scholar] [CrossRef] [PubMed]
- Takeda, M.; Leser, G.P.; Russell, C.J.; Lamb, R.A. Influenza virus hemagglutinin concentrates in lipid raft microdomains for efficient viral fusion. Proc. Natl. Acad. Sci. USA 2003, 100, 14610–14617. [Google Scholar] [CrossRef] [PubMed]
- Ma, K.; Wang, Y.J.; Wang, J.F. Influenza virus assembly and budding in lipid rafts. Prog. Biochem. Biophys. 2015, 42, 495–500. [Google Scholar]
- Rossman, J.S.; Jing, X.H.; Leser, G.P.; Lamb, R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell 2010, 142, 902–913. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Turpin, E.A.; Bultmann, H.; Brandt, C.R.; Schultz-Cherry, S. Inhibition of influenza virus infection by a novel antiviral peptide that targets viral attachment to cells. J. Virol. 2006, 80, 11960–11967. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Settles, E.W.; Brandt, C.R.; Schultz-Cherry, S. Identification of the minimal active sequence of an anti-influenza virus peptide. Antimicrob. Agents Chemother. 2011, 55, 1810–1813. [Google Scholar] [CrossRef] [PubMed]
- Rajik, M.; Jahanshiri, F.; Omar, A.R.; Ideris, A.; Hassan, S.S.; Yusoff, K. Identification and characterisation of a novel anti-viral peptide against avian influenza virus H9N2. Virol. J. 2009, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nicol, M.Q.; Ligertwood, Y.; Bacon, M.N.; Dutia, B.M.; Nash, A.A. A novel family of peptides with potent activity against influenza A viruses. J. Gen. Virol. 2012, 93, 980–986. [Google Scholar] [CrossRef] [PubMed]
- Matsubara, T.; Onishi, A.; Saito, T.; Shimada, A.; Inoue, H.; Taki, T.; Nagata, K.; Okahata, Y.; Sato, T. Sialic acid-mimic peptides as hemagglutinin inhibitors for anti-influenza therapy. J. Med. Chem. 2010, 53, 4441–4449. [Google Scholar] [CrossRef] [PubMed]
- Huttl, C.; Hettrich, C.; Miller, R.; Paulke, B.R.; Henklein, P.; Rawel, H.; Bier, F.F. Self-assembled peptide amphiphiles function as multivalent binder with increased hemagglutinin affinity. BMC Biotechnol. 2013, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Cederlund, A.; Gudmundsson, G.H.; Agerberth, B. Antimicrobial peptides important in innate immunity. FEBS J. 2011, 278, 3942–3951. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, J.; Schneider, C.; Heinbockel, L.; Brandenburg, K.; Reimer, R.; Gabriel, G. A new class of synthetic anti-lipopolysaccharide peptides inhibits influenza A virus replication by blocking cellular attachment. Antivir. Res. 2014, 104, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Doss, M.; White, M.R.; Tecle, T.; Gantz, D.; Crouch, E.C.; Jung, G.; Ruchala, P.; Waring, A.J.; Lehrer, R.I.; Hartshorn, K.L. Interactions of α-, β-, and θ-defensins with influenza A virus and surfactant protein D. J. Immunol. 2009, 182, 7878–7887. [Google Scholar] [CrossRef] [PubMed]
- Ammendolia, M.G.; Agamennone, M.; Pietrantoni, A.; Lannutti, F.; Siciliano, R.A.; de Giulio, B.; Amici, C.; Superti, F. Bovine lactoferrin-derived peptides as novel broad-spectrum inhibitors of influenza virus. Pathog. Glob. Health 2012, 106, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.L.; Zhao, Z.H.; Zhou, D.H.; Chen, Y.Q.; Hong, W.; Cao, L.Y.; Yang, J.Y.; Zhang, Y.; Shi, W.; Cao, Z.J.; et al. Virucidal activity of a scorpion venom peptide variant mucroporin-M1 against measles, SARS-CoV and influenza H5N1 viruses. Peptides 2011, 32, 1518–1525. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, S.; Tecle, T.; Verma, A.; Crouch, E.; White, M.; Hartshorn, K.L. The human cathelicidin LL-37 inhibits influenza A viruses through a mechanism distinct from that of surfactant protein D or defensins. J. Gen. Virol. 2013, 94, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Ghanem, A.; Mayer, D.; Chase, G.; Tegge, W.; Frank, R.; Kochs, G.; Garcia-Sastre, A.; Schwemmle, M. Peptide-mediated interference with influenza A virus polymerase. J. Virol. 2007, 81, 7801–7804. [Google Scholar] [CrossRef] [PubMed]
- Chase, G.; Wunderlich, K.; Reuther, P.; Schwemmle, M. Identification of influenza virus inhibitors which disrupt of viral polymerase protein-protein interactions. Methods 2011, 55, 188–191. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Li, M.M.; Shen, X.T.; Liu, S.W. Influenza A virus entry inhibitors targeting the hemagglutinin. Viruses 2013, 5, 352–373. [Google Scholar] [CrossRef] [PubMed]
- Wunderlich, K.; Mayer, D.; Ranadheera, C.; Holler, A.S.; Manz, B.; Martin, A.; Chase, G.; Tegge, W.; Frank, R.; Kessler, U.; et al. Identification of a PA-binding peptide with inhibitory activity against influenza A and B virus replication. PLoS ONE 2009, 4, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Conti, G.; Magliani, W.; Conti, S.; Nencioni, L.; Sgarbanti, R.; Palamara, A.T.; Polonelli, L. Therapeutic activity of an anti-idiotypic antibody-derived killer peptide against influenza A virus experimental infection. Antimicrob. Agents Chemother. 2008, 52, 4331–4337. [Google Scholar] [CrossRef] [PubMed]
- Salvatore, M.; Garcia-Sastre, A.; Ruchala, P.; Lehrer, R.I.; Chang, T.; Klotman, M.E. α-Defensin inhibits influenza virus replication by cell-mediated mechanism(s). J. Infect. Dis. 2007, 196, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Judd, A.K.; Sanchez, A.; Bucher, D.J.; Huffman, J.H.; Bailey, K.; Sidwell, R.W. In vivo anti-influenza virus activity of a zinc finger peptide. Antimicrob. Agents Chemother. 1997, 41, 687–692. [Google Scholar] [PubMed]
- Nasser, E.H.; Judd, A.K.; Sanchez, A.; Anastasiou, D.; Bucher, D.J. Antiviral activity of influenza virus M1 zinc finger peptides. J. Virol. 1996, 70, 8639–8644. [Google Scholar] [PubMed]
- Ahmed, C.M.; Dabelic, R.; Waiboci, L.W.; Jager, L.D.; Heron, L.L.; Johnson, H.M. SOCS-1 mimetics protect mice against lethal poxvirus infection: Identification of a novel endogenous antiviral system. J. Virol. 2009, 83, 1402–1415. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.P.; Chu, Y.H. Using surface plasmon resonance to directly determine binding affinities of combinatorially selected cyclopeptides and their linear analogs to a streptavidin chip. Anal. Biochem. 2005, 340, 74–79. [Google Scholar] [CrossRef] [PubMed]
- Udugamasooriya, D.G.; Spaller, M.R. Conformational constraint in protein ligand design and the inconsistency of binding entropy. Biopolymers 2008, 89, 653–667. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Cole, A.M.; Hong, T.; Waring, A.J.; Lehrer, R.I. Retrocyclin, an antiretroviral θ-defensin, is a lectin. J. Immunol. 2003, 170, 4708–4716. [Google Scholar] [CrossRef] [PubMed]
- Doss, M.; White, M.R.; Tecle, T.; Hartshorn, K.L. Human defensins and LL-37 in mucosal immunity. J. Leukoc. Biol. 2010, 87, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Gutsmann, T.; Razquin-Olazaran, I.; Kowalski, I.; Kaconis, Y.; Howe, J.; Bartels, R.; Hornef, M.; Schurholz, T.; Rossle, M.; Sanchez-Gomez, S.; et al. New antiseptic peptides to protect against endotoxin-mediated shock. Antimicrob. Agents Chemother. 2010, 54, 3817–3824. [Google Scholar] [CrossRef] [PubMed]
- Krepstakies, M.; Lucifora, J.; Nagel, C.H.; Zeisel, M.B.; Holstermann, B.; Hohenberg, H.; Kowalski, I.; Gutsmann, T.; Baumert, T.F.; Brandenburg, K.; et al. A new class of synthetic peptide inhibitors blocks attachment and entry of human pathogenic viruses. J. Infect. Dis. 2012, 205, 1654–1664. [Google Scholar] [CrossRef] [PubMed]
- Needham, B.D.; Trent, M.S. Fortifying the barrier: The impact of lipid a remodelling on bacterial pathogenesis. Nat. Rev. Microbiol. 2013, 11, 467–481. [Google Scholar] [CrossRef] [PubMed]
- Anaya-Lopez, J.L.; Lopez-Meza, J.E.; Ochoa-Zarzosa, A. Bacterial resistance to cationic antimicrobial peptides. Crit. Rev. Microbiol. 2013, 39, 180–195. [Google Scholar] [CrossRef] [PubMed]
- Mercer, D.K.; O’Neil, D.A. Peptides as the next generation of anti-infectives. Future Med. Chem. 2013, 5, 315–337. [Google Scholar] [CrossRef] [PubMed]
- Marcos, J.F.; Gandia, M. Antimicrobial peptides: To membranes and beyond. Expert Opin. Drug Discov. 2009, 4, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, L.; Knoelker, H.-J.; Simons, K. Subcellular targeting strategies for drug design and delivery. Nat. Rev. Drug Discov. 2010, 9, 29–42. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.-Y.G.; Mookherjee, N. Multiple immune-modulatory functions of cathelicidin host defense peptides. Front. Immun. 2012, 3, 149. [Google Scholar] [CrossRef] [PubMed]
- Barlow, P.G.; Svoboda, P.; Mackellar, A.; Nash, A.A.; York, I.A.; Pohl, J.; Davidson, D.J.; Donis, R.O. Antiviral activity and increased host defense against influenza infection elicited by the human cathelicidin LL-37. PLoS ONE 2011, 6, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dean, R.E.; O’Brien, L.M.; Thwaite, J.E.; Fox, M.A.; Atkins, H.; Ulaeto, D.O. A carpet-based mechanism for direct antimicrobial peptide activity against vaccinia virus membranes. Peptides 2010, 31, 1966–1972. [Google Scholar] [PubMed]
- Pietrantoni, A.; Di Biase, A.M.; Tinari, A.; Marchetti, M.; Valenti, P.; Seganti, L.; Superti, F. Bovine lactoferrin inhibits adenovirus infection by interacting with viral structural polypeptides. Antimicrob. Agents Chemother. 2003, 47, 2688–2691. [Google Scholar] [PubMed]
- Pietrantoni, A.; Dofrelli, E.; Tinari, A.; Ammendolia, M.G.; Puzelli, S.; Fabiani, C.; Donatelli, I.; Superti, F. Bovine lactoferrin inhibits influenza A virus induced programmed cell death in vitro. Biometals 2010, 23, 465–475. [Google Scholar] [CrossRef] [PubMed]
- Balcao, V.M.; Costa, C.I.; Matos, C.M.; Moutinho, C.G.; Amorim, M.; Pintado, M.E.; Gomes, A.P.; Vila, M.M.; Teixeira, J.A. Nanoencapsulation of bovine lactoferrin for food and biopharmaceutical applications. Food Hydrocoll. 2013, 32, 425–431. [Google Scholar] [CrossRef] [Green Version]
- Bibi, S.; Lattmann, E.; Mohammed, A.R.; Perrie, Y. Trigger release liposome systems: Local and remote controlled delivery? J. Microencapsul. 2012, 29, 262–276. [Google Scholar] [CrossRef] [PubMed]
- Raghuraman, H.; Chattopadhyay, A. Melittin: A membrane-active peptide with diverse functions. Biosci. Rep. 2007, 27, 189–223. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.T.; Hung, W.C.; Chen, F.Y.; Huang, H.W. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 5087–5092. [Google Scholar] [CrossRef] [PubMed]
- Ladokhin, A.S.; White, S.H. ‘Detergent-like’ permeabilization of anionic lipid vesicles by melittin. Biochim. Biophys. Acta 2001, 1514, 253–260. [Google Scholar] [CrossRef]
- Zhang, N.Z.; Qi, J.X.; Feng, S.J.; Gao, F.; Liu, J.; Pan, X.C.; Chen, R.; Li, Q.R.; Chen, Z.S.; Li, X.Y.; et al. Crystal structure of swine major histocompatibility complex class I SLA-1*0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic t lymphocyte epitope peptides. J. Virol. 2011, 85, 11709–11724. [Google Scholar] [CrossRef] [PubMed]
- Gordon-Grossman, M.; Zimmermann, H.; Wolf, S.G.; Shai, Y.; Goldfarb, D. Investigation of model membrane disruption mechanism by melittin using pulse electron paramagnetic resonance spectroscopy and cryogenic transmission electron microscopy. J. Phys. Chem. B 2012, 116, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Gordon-Grossman, M.; Gofman, Y.; Zimmermann, H.; Frydman, V.; Shai, Y.; Ben-Tal, N.; Goldfarb, D. A combined pulse EPR and monte carlo simulation study provides molecular insight on peptide-membrane interactions. J. Phys. Chem. B 2009, 113, 15128. [Google Scholar] [CrossRef]
- Oren, Z.; Shai, Y. Mode of action of linear amphipathic α-helical antimicrobial peptides. Biopolymers 1998, 47, 451–463. [Google Scholar] [CrossRef]
- Benachir, T.; Lafleur, M. Study of vesicle leakage induced by melittin. Biochim. Biophys. Acta 1995, 1235, 452–460. [Google Scholar] [CrossRef]
- Lu, N.Y.; Yang, K.; Li, J.L.; Yuan, B.; Ma, Y.Q. Vesicle deposition and subsequent membrane-melittin interactions on different substrates: A QCM-D experiment. Biochim. Biophys. Acta 2013, 1828, 1918–1925. [Google Scholar] [CrossRef] [PubMed]
- Li, W.Y.; Yang, X.F.; Jiang, Y.; Wang, B.N.; Yang, Y.; Jiang, Z.H.; Li, M.Y. Inhibition of influenza A virus replication by RNA interference targeted against the PB1 subunit of the RNA polymerase gene. Arch. Virol. 2011, 156, 1979–1987. [Google Scholar] [CrossRef] [PubMed]
- Huet, S.; Avilov, S.V.; Ferbitz, L.; Daigle, N.; Cusack, S.; Ellenberg, J. Nuclear import and assembly of influenza A virus RNA polymerase studied in live cells by fluorescence cross-correlation spectroscopy. J. Virol. 2010, 84, 1254–1264. [Google Scholar] [CrossRef] [PubMed]
- Manz, B.; Gotz, V.; Wunderlich, K.; Eisel, J.; Kirchmair, J.; Stech, J.; Stech, O.; Chase, G.; Frank, R.; Schwemmle, M. Disruption of the viral polymerase complex assembly as a novel approach to attenuate influenza A virus. J. Biol. Chem. 2011, 286, 8414–8424. [Google Scholar] [CrossRef] [PubMed]
- Ruigrok, R.W.H.; Crepin, T.; Hart, D.J.; Cusack, S. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 2010, 20, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Engler, A.C.; Shukla, A.; Puranam, S.; Buss, H.G.; Jreige, N.; Hammond, P.T. Effects of side group functionality and molecular weight on the activity of synthetic antimicrobial polypeptides. Biomacromolecules 2011, 12, 1666–1674. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.Q.; Shan, A.S.; Dong, N.; Gu, Y.; Sun, W.Y.; Hu, W.N.; Feng, X.J. Cell selectivity and interaction with model membranes of Val/Arg-rich peptides. J. Pept. Sci. 2011, 17, 520–526. [Google Scholar] [CrossRef] [PubMed]
- Fechter, P.; Mingay, L.; Sharps, J.; Chambers, A.; Fodor, E.; Brownlee, G.G. Two aromatic residues in the PB2 subunit of influenza A RNA polymerase are crucial for cap binding. J. Biol. Chem. 2003, 278, 20381–20388. [Google Scholar] [CrossRef] [PubMed]
- Dias, A.; Bouvier, D.; Crepin, T.; McCarthy, A.A.; Hart, D.J.; Baudin, F.; Cusack, S.; Ruigrok, R.W.H. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 2009, 458, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.W.; Bartlam, M.; Lou, Z.Y.; Chen, S.D.; Zhou, J.; He, X.J.; Lv, Z.Y.; Ge, R.W.; Li, X.M.; Deng, T.; et al. Crystal structure of an avian influenza polymerase PA(n) reveals an endonuclease active site. Nature 2009, 458, 909–913. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.Y.; Yu, W.L.; McBride, R.; Li, Y.; Chen, L.M.; Donis, R.O.; Tong, S.X.; Paulson, J.C.; Wilson, I.A. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc. Natl. Acad. Sci. USA 2013, 110, 1458–1463. [Google Scholar] [CrossRef] [PubMed]
- Stephens, C.; Kazan, K.; Goulter, K.C.; Maclean, D.J.; Manners, J.M. The mode of action of the plant antimicrobial peptide MiAMP1 differs from that of its structural homologue, the yeast killer toxin WmKT. FEMS Microbiol. Lett. 2005, 243, 205–210. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.S.; Wiens, M.E.; Smith, J.G. Antiviral mechanisms of human defensins. J. Mol. Biol. 2013, 425, 4965–4980. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Chang, T.L. Defensins in viral infection. In Small Wonders: Peptides for Disease Control; American Chemical Society: Washington, DC, USA, 2012; pp. 137–171. [Google Scholar]
- Mohan, T.; Sharma, C.; Bhat, A.A.; Rao, D.N. Modulation of HIV peptide antigen specific cellular immune response by synthetic α- and β-defensin peptides. Vaccine 2013, 31, 1707–1716. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skalickova, S.; Heger, Z.; Krejcova, L.; Pekarik, V.; Bastl, K.; Janda, J.; Kostolansky, F.; Vareckova, E.; Zitka, O.; Adam, V.; et al. Perspective of Use of Antiviral Peptides against Influenza Virus. Viruses 2015, 7, 5428-5442. https://doi.org/10.3390/v7102883
Skalickova S, Heger Z, Krejcova L, Pekarik V, Bastl K, Janda J, Kostolansky F, Vareckova E, Zitka O, Adam V, et al. Perspective of Use of Antiviral Peptides against Influenza Virus. Viruses. 2015; 7(10):5428-5442. https://doi.org/10.3390/v7102883
Chicago/Turabian StyleSkalickova, Sylvie, Zbynek Heger, Ludmila Krejcova, Vladimir Pekarik, Karel Bastl, Jozef Janda, Frantisek Kostolansky, Eva Vareckova, Ondrej Zitka, Vojtech Adam, and et al. 2015. "Perspective of Use of Antiviral Peptides against Influenza Virus" Viruses 7, no. 10: 5428-5442. https://doi.org/10.3390/v7102883