3D Analysis of HCMV Induced-Nuclear Membrane Structures by FIB/SEM Tomography: Insight into an Unprecedented Membrane Morphology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Virus
2.2. Sample Preparation for FIB/SEM Tomography and Transmission Electron Microscopy (TEM)
2.3. FIB/SEM Tomography

2.4. Data Processing
2.5. (Serial-)Ultrathin Sectioning and TEM
3. Results
3.1. FIB/SEM Tomography is a Powerful Tool for Virological Research

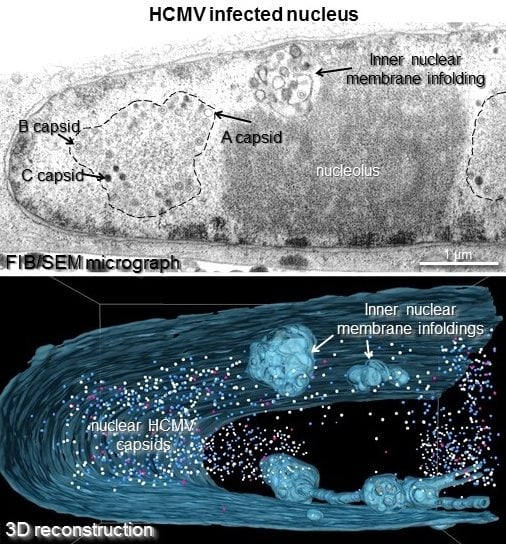
3.2. Three Dimensional FIB/SEM Tomography Reveals the Enormous Structural Changes to the Inner Nuclear Membrane in HCMV Infected Cells
3.3. 1st, 2nd and 3rd Order Infoldings






3.4. Three-Dimensional Distribution of Nuclear HCMV Capsids
| Number of Capsids (% of Total Capsids) | Distribution (%) | ||||
|---|---|---|---|---|---|
| A Capsids | B Capsids | C Capsids | |||
| Total Capsids | 5498 | (100.0%) | 6.0 | 52.0 | 42.0 |
| -Volume 1 | 4160 | (75.7%) | 6.0 | 52.9 | 41.1 |
| -Volume 2 | 1338 | (24.3%) | 5.9 | 49.3 | 44.8 |
| In Nucleoplasm | |||||
| (Non-Enveloped) | 5370 | (97.7%) | 6.0 | 51.8 | 42.2 |
| In Infoldings | |||||
| - Enveloped in 1st Order Infoldings | 45 | (0.8%) | 0.0 | 57.8 | 42.2 |
| - Non-Enveloped in 2nd Order Infoldings | 83 | (1.5%) | 6.0 | 66.3 | 27.7 |
3.5. Quantification of Capsids in Infoldings
4. Discussion
4.1. Three-Dimensional Imaging Reveals a Hierarchical Infolding Structure
4.2. The Pushing Membrane Model
4.3. Three-Dimensional Analysis of Nuclear HCMV Capsids
4.4. Function of the Nuclear Infoldings
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Buser, C.; Walther, P.; Mertens, T.; Michel, D. Cytomegalovirus Primary Envelopment Occurs at Large Infoldings of the Inner Nuclear Membrane. J. Virol. 2007, 81, 3042–3048. [Google Scholar] [CrossRef] [PubMed]
- Homman-Loudiyi, M.; Hultenby, K.; Britt, W.; Soderberg-Naucler, C. Envelopment of Human Cytomegalovirus Occurs by Budding into Golgi-Derived Vacuole Compartments Positive for gB, Rab 3, Trans-Golgi Network 46, and Mannosidase II. J. Virol. 2003, 77, 3191–3203. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.J.; Adler, B.; Sampaio, K.L.; Digel, M.; Jahn, G.; Ettischer, N.; Stierhof, Y.-D.; Scrivano, L.; Koszinowski, U.; Mach, M.; et al. UL74 of human cytomegalovirus contributes to virus release by promoting secondary envelopment of virions. J. Virol. 2008, 82, 2802–2812. [Google Scholar] [CrossRef] [PubMed]
- Meissner, C.S.; Suffner, S.; Schauflinger, M.; von Einem, J.; Bogner, E. A Leucine Zipper Motif of a Tegument Protein Triggers Final Envelopment of Human Cytomegalovirus. J. Virol. 2011, 86, 3370–3382. [Google Scholar] [CrossRef] [PubMed]
- Schauflinger, M.; Fischer, D.; Schreiber, A.; Chevillotte, M.; Walther, P.; Mertens, T.; von Einem, J. The Tegument Protein UL71 of Human Cytomegalovirus Is Involved in Late Envelopment and Affects Multivesicular Bodies. J. Virol. 2011, 85, 3821–3832. [Google Scholar] [CrossRef] [PubMed]
- Seo, J.-Y.; Britt, W.J. Cytoplasmic Envelopment of Human Cytomegalovirus Requires the Postlocalization Function of Tegument Protein pp28 within the Assembly Compartment. J. Virol. 2007, 81, 6536–6547. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Loveland, A.N.; Kattenhorn, L.M.; Ploegh, H.L.; Gibson, W. High-Molecular-Weight Protein (pUL48) of Human Cytomegalovirus Is a Competent Deubiquitinating Protease: Mutant Viruses Altered in Its Active-Site Cysteine or Histidine Are Viable. J. Virol. 2006, 80, 6003–6012. [Google Scholar] [CrossRef] [PubMed]
- Gibson, W. Structure and assembly of the virion. Intervirology 1996, 39, 389–400. [Google Scholar] [PubMed]
- Gibson, W. Structure and formation of the cytomegalovirus virion. Curr. Top. Microbiol. Immunol. 2008, 325, 187–204. [Google Scholar] [PubMed]
- Varnum, S.M.; Streblow, D.N.; Monroe, M.E.; Smith, P.; Auberry, K.J.; Pasa-Tolic, L.; Wang, D.; Camp, D.G.; Rodland, K.; Wiley, S.; et al. Identification of Proteins in Human Cytomegalovirus (HCMV) Particles: The HCMV Proteome. J. Virol. 2004, 78, 10960–10966. [Google Scholar] [CrossRef] [PubMed]
- Schmolke, S.; Kern, H.F.; Drescher, P.; Jahn, G.; Plachter, B. The dominant phosphoprotein pp65 (UL83) of human cytomegalovirus is dispensable for growth in cell culture. J. Virol. 1995, 69, 5959–5968. [Google Scholar] [PubMed]
- Tandon, R.; Mocarski, E.S.; Conway, J.F. The A, B, Cs of Herpesvirus Capsids. Viruses 2015, 7, 899–914. [Google Scholar] [CrossRef] [PubMed]
- Toropova, K.; Huffman, J.B.; Homa, F.L.; Conway, J.F. The herpes simplex virus 1 UL17 protein is the second constituent of the capsid vertex-specific component required for DNA packaging and retention. J. Virol. 2011, 85, 7513–7522. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Trang, P.; Shah, S.; Atanasov, I.; Kim, Y.-H.; Bai, Y.; Zhou, Z.H.; Liu, F. Dissecting human cytomegalovirus gene function and capsid maturation by ribozyme targeting and electron cryomicroscopy. Proc. Natl. Acad. Sci. USA 2005, 102, 7103–7108. [Google Scholar] [CrossRef] [PubMed]
- Panté, N.; Kann, M. Nuclear Pore Complex Is Able to Transport Macromolecules with Diameters of ∼39 nm. Mol. Biol. Cell 2002, 13, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Leuzinger, H.; Ziegler, U.; Schraner, E.M.; Fraefel, C.; Glauser, D.L.; Heid, I.; Ackermann, M.; Mueller, M.; Wild, P. Herpes Simplex Virus 1 Envelopment Follows Two Diverse Pathways. J. Virol. 2005, 79, 13047–13059. [Google Scholar] [CrossRef] [PubMed]
- Skepper, J.N.; Whiteley, A.; Browne, H.; Minson, A. Herpes Simplex Virus Nucleocapsids Mature to Progeny Virions by an Envelopment → Deenvelopment → Reenvelopment Pathway. J. Virol. 2001, 75, 5697–5702. [Google Scholar] [CrossRef] [PubMed]
- Stackpole, C.W. Herpes-Type Virus of the Frog Renal Adenocarcinoma I. Virus Development in Tumor Transplants Maintained at Low Temperature. J. Virol. 1969, 4, 75–93. [Google Scholar] [PubMed]
- Mettenleiter, T.C.; Müller, F.; Granzow, H.; Klupp, B.G. The way out: What we know and do not know about herpesvirus nuclear egress. Cell. Microbiol. 2013, 15, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Schulz, K.S.; Klupp, B.G.; Granzow, H.; Paßvogel, L.; Mettenleiter, T.C. Herpesvirus nuclear egress: Pseudorabies Virus can simultaneously induce nuclear envelope breakdown and exit the nucleus via the envelopment–deenvelopment-pathway. Virus Res. 2015, 209, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.; Chou, C.; Li, H.; Hai, R.; Patterson, D.; Stolc, V.; Zhu, H.; Liu, F. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. 2003, 100, 14223–14228. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Silva, M.C.; Shenk, T. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. 2003, 100, 12396–12401. [Google Scholar] [CrossRef] [PubMed]
- Marschall, M.; Marzi, A.; aus dem Siepen, P.; Jochmann, R.; Kalmer, M.; Auerochs, S.; Lischka, P.; Leis, M.; Stamminger, T. Cellular p32 Recruits Cytomegalovirus Kinase pUL97 to Redistribute the Nuclear Lamina. J. Biol. Chem. 2005, 280, 33357–33367. [Google Scholar] [CrossRef] [PubMed]
- Muranyi, W.; Haas, J.; Wagner, M.; Krohne, G.; Koszinowski, U.H. Cytomegalovirus Recruitment of Cellular Kinases to Dissolve the Nuclear Lamina. Science 2002, 297, 854–857. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Kamil, J.P.; Coughlin, M.; Reim, N.I.; Coen, D.M. Human Cytomegalovirus UL50 and UL53 Recruit Viral Protein Kinase UL97, not Protein Kinase C, for Disruption of Nuclear Lamina and Nuclear Egress in Infected Cells. J. Virol. 2014, 88, 249–262. [Google Scholar] [CrossRef] [PubMed]
- Hatch, E.M.; Hetzer, M.W. RNP Export by Nuclear Envelope Budding. Cell 2012, 149, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Speese, S.D.; Ashley, J.; Jokhi, V.; Nunnari, J.; Barria, R.; Li, Y.; Ataman, B.; Koon, A.; Chang, Y.-T.; Li, Q.; et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 2012, 149, 832–846. [Google Scholar] [CrossRef] [PubMed]
- Bayer, C.; Varani, S.; Wang, L.; Walther, P.; Zhou, S.; Straschewski, S.; Bachem, M.; Söderberg-Naucler, C.; Mertens, T.; Frascaroli, G. Human Cytomegalovirus Infection of M1 and M2 Macrophages Triggers Inflammation and Autologous T-Cell Proliferation. J. Virol. 2013, 87, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Malhas, A.; Goulbourne, C.; Vaux, D.J. The nucleoplasmic reticulum: form and function. Trends Cell Biol. 2011, 21, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Ballerini, M.; Milani, M.; Costato, M.; Squadrini, F.; Turcu, I.C. Life science applications of focused ion beams (FIB). Eur. J. Histochem. EJH 1997, 41 (Suppl. 2), 89–90. [Google Scholar] [PubMed]
- Becker, C.; Ali, K.; Knott, G.; Fua, P. Learning context cues for synapse segmentation. IEEE Trans. Med. Imaging 2013, 32, 1864–1877. [Google Scholar] [CrossRef] [PubMed]
- Hekking, L.H.P.; Lebbink, M.N.; de Winter, D.A.M.; Schneijdenberg, C.T.W.M.; Brand, C.M.; Humbel, B.M.; Verkleij, A.J.; Post, J.A. Focused ion beam-scanning electron microscope: Exploring large volumes of atherosclerotic tissue. J. Microsc. 2009, 235, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Knott, G.; Rosset, S.; Cantoni, M. Focussed ion beam milling and scanning electron microscopy of brain tissue. J. Vis. Exp. JoVE 2011, e2588. [Google Scholar] [CrossRef] [PubMed]
- Leser, V.; Drobne, D.; Pipan, Z.; Milani, M.; Tatti, F. Comparison of different preparation methods of biological samples for FIB milling and SEM investigation. J. Microsc. 2009, 233, 309–319. [Google Scholar] [CrossRef] [PubMed]
- Schertel, A.; Snaidero, N.; Han, H.-M.; Ruhwedel, T.; Laue, M.; Grabenbauer, M.; Möbius, W. Cryo FIB-SEM: Volume imaging of cellular ultrastructure in native frozen specimens. J. Struct. Biol. 2013, 184, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Schroeder-Reiter, E.; Pérez-Willard, F.; Zeile, U.; Wanner, G. Focused ion beam (FIB) combined with high resolution scanning electron microscopy: a promising tool for 3D analysis of chromosome architecture. J. Struct. Biol. 2009, 165, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Kizilyaprak, C.; Daraspe, J.; Humbel, B. Focused ion beam scanning electron microscopy in biology. J. Microsc. 2014, 254, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Villinger, C.; Gregorius, H.; Kranz, C.; Höhn, K.; Münzberg, C.; Wichert, G.; Mizaikoff, B.; Wanner, G.; Walther, P. FIB/SEM tomography with TEM-like resolution for 3D imaging of high-pressure frozen cells. Histochem. Cell Biol. 2012, 138, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Villinger, C.; Schauflinger, M.; Gregorius, H.; Kranz, C.; Höhn, K.; Nafeey, S.; Walther, P. Three-dimensional imaging of adherent cells using FIB/SEM and STEM. Methods Mol. Biol. Clifton NJ 2014, 1117, 617–638. [Google Scholar]
- Sinzger, C.; Hahn, G.; Digel, M.; Katona, R.; Sampaio, K.L.; Messerle, M.; Hengel, H.; Koszinowski, U.; Brune, W.; Adler, B. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 2008, 89, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Schauflinger, M.; Villinger, C.; Mertens, T.; Walther, P.; von Einem, J. Analysis of human cytomegalovirus secondary envelopment by advanced electron microscopy: HCMV morphogenesis. Cell. Microbiol. 2013, 15, 305–314. [Google Scholar] [CrossRef] [PubMed]
- Walther, P.; Ziegler, A. Freeze substitution of high-pressure frozen samples: the visibility of biological membranes is improved when the substitution medium contains water. J. Microsc. 2002, 208, 3–10. [Google Scholar] [CrossRef] [PubMed]
- ImageJ. Availiable online: http://rsbweb.nih.gov/ij/index.html (accessed on 27 October 2015).
- Kremer, J.R.; Mastronarde, D.N.; McIntosh, J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996, 116, 71–76. [Google Scholar] [CrossRef] [PubMed]
- The IMOD Home Page. http://bio3d.colorado.edu/imod/ (accessed on 27 October 2015).
- Strang, B.L. Viral and cellular subnuclear structures in human cytomegalovirus-infected cells. J. Gen. Virol. 2015, 96, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Klupp, B.G.; Granzow, H.; Mettenleiter, T.C. Nuclear Envelope Breakdown Can Substitute for Primary Envelopment-Mediated Nuclear Egress of Herpesviruses. J. Virol. 2011, 85, 8285–8292. [Google Scholar] [CrossRef] [PubMed]
- Hagen, C.; Guttmann, P.; Klupp, B.; Werner, S.; Rehbein, S.; Mettenleiter, T.C.; Schneider, G.; Grunewald, K. Correlative VIS-fluorescence and soft X-ray cryo-microscopy/tomography of adherent cells. J. Struct. Biol. 2012, 177, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Klupp, B.G.; Granzow, H.; Fuchs, W.; Keil, G.M.; Finke, S.; Mettenleiter, T.C. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 7241–7246. [Google Scholar] [CrossRef] [PubMed]
- Pignatelli, S.; Monte, P.D.; Landini, M.P.; Severi, B.; Nassiri, R.; Gilloteaux, J.; Papadimitriou, J.M.; Shellam, G.R. Cytomegalovirus Primary Envelopment at Large Nuclear Membrane Infoldings: What’s New? J. Virol. 2007, 81, 7320–7322. [Google Scholar] [CrossRef] [PubMed]
- Prüfert, K.; Vogel, A.; Krohne, G. The lamin CxxM motif promotes nuclear membrane growth. J. Cell Sci. 2004, 117, 6105–6116. [Google Scholar] [CrossRef] [PubMed]
- Roffman, E.; Albert, J.P.; Goff, J.P.; Frenkel, N. Putative site for the acquisition of human herpesvirus 6 virion tegument. J. Virol. 1990, 64, 6308–6313. [Google Scholar] [PubMed]
- Sutter, E.; de Oliveira, A.P.; Tobler, K.; Schraner, E.M.; Sonda, S.; Kaech, A.; Lucas, M.S.; Ackermann, M.; Wild, P. Herpes simplex virus 1 induces de novo phospholipid synthesis. Virology 2012, 429, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Wild, P.; Engels, M.; Senn, C.; Tobler, K.; Ziegler, U.; Schraner, E.M.; Loepfe, E.; Ackermann, M.; Mueller, M.; Walther, P. Impairment of Nuclear Pores in Bovine Herpesvirus 1-Infected MDBK Cells. J. Virol. 2005, 79, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Tandon, R.; Mocarski, E.S. Viral and host control of cytomegalovirus maturation. Trends Microbiol. 2012, 20, 392–401. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villinger, C.; Neusser, G.; Kranz, C.; Walther, P.; Mertens, T. 3D Analysis of HCMV Induced-Nuclear Membrane Structures by FIB/SEM Tomography: Insight into an Unprecedented Membrane Morphology. Viruses 2015, 7, 5686-5704. https://doi.org/10.3390/v7112900
Villinger C, Neusser G, Kranz C, Walther P, Mertens T. 3D Analysis of HCMV Induced-Nuclear Membrane Structures by FIB/SEM Tomography: Insight into an Unprecedented Membrane Morphology. Viruses. 2015; 7(11):5686-5704. https://doi.org/10.3390/v7112900
Chicago/Turabian StyleVillinger, Clarissa, Gregor Neusser, Christine Kranz, Paul Walther, and Thomas Mertens. 2015. "3D Analysis of HCMV Induced-Nuclear Membrane Structures by FIB/SEM Tomography: Insight into an Unprecedented Membrane Morphology" Viruses 7, no. 11: 5686-5704. https://doi.org/10.3390/v7112900
APA StyleVillinger, C., Neusser, G., Kranz, C., Walther, P., & Mertens, T. (2015). 3D Analysis of HCMV Induced-Nuclear Membrane Structures by FIB/SEM Tomography: Insight into an Unprecedented Membrane Morphology. Viruses, 7(11), 5686-5704. https://doi.org/10.3390/v7112900