Use of Cellular Decapping Activators by Positive-Strand RNA Viruses
Abstract
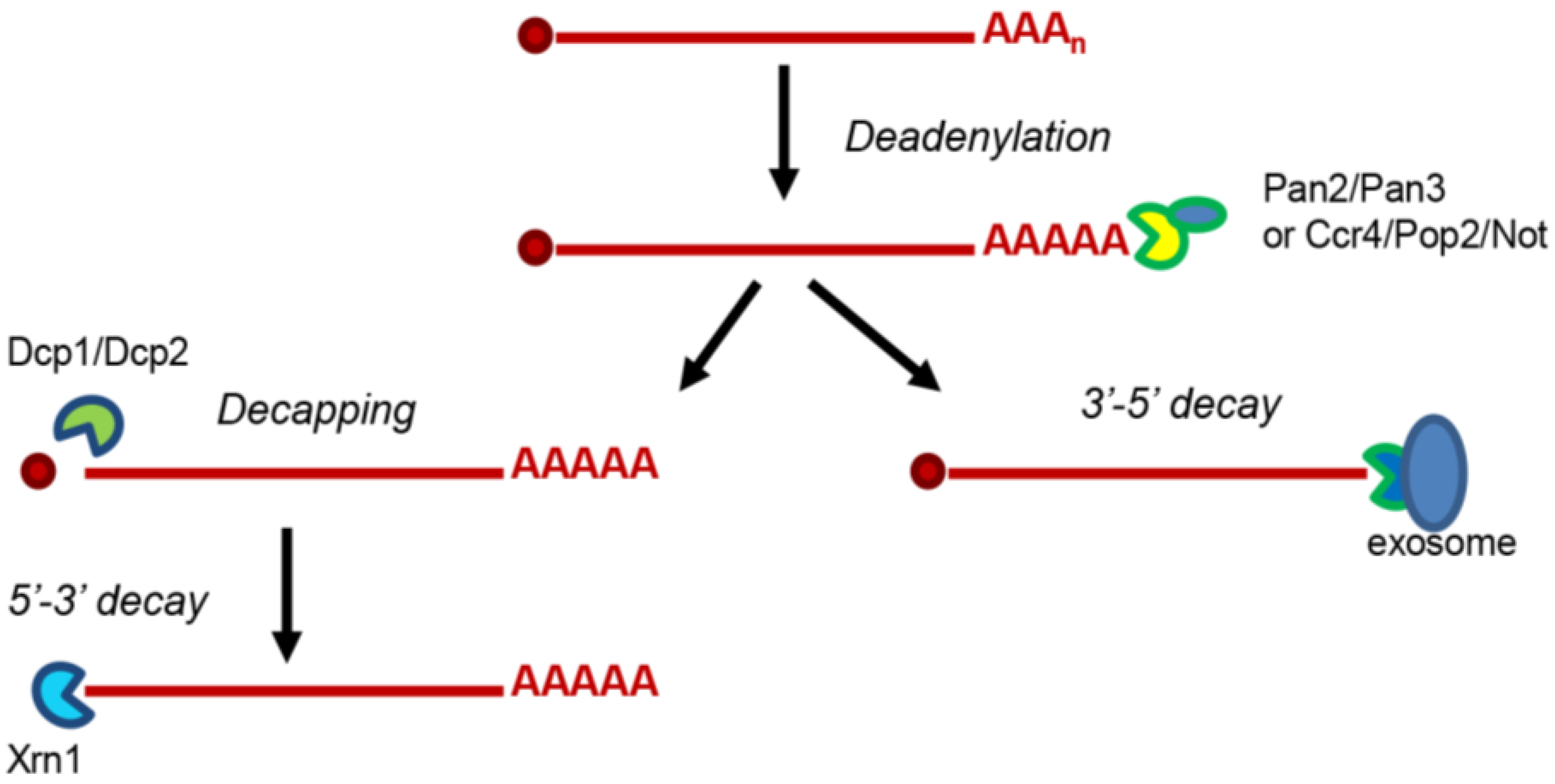
:1. Introduction
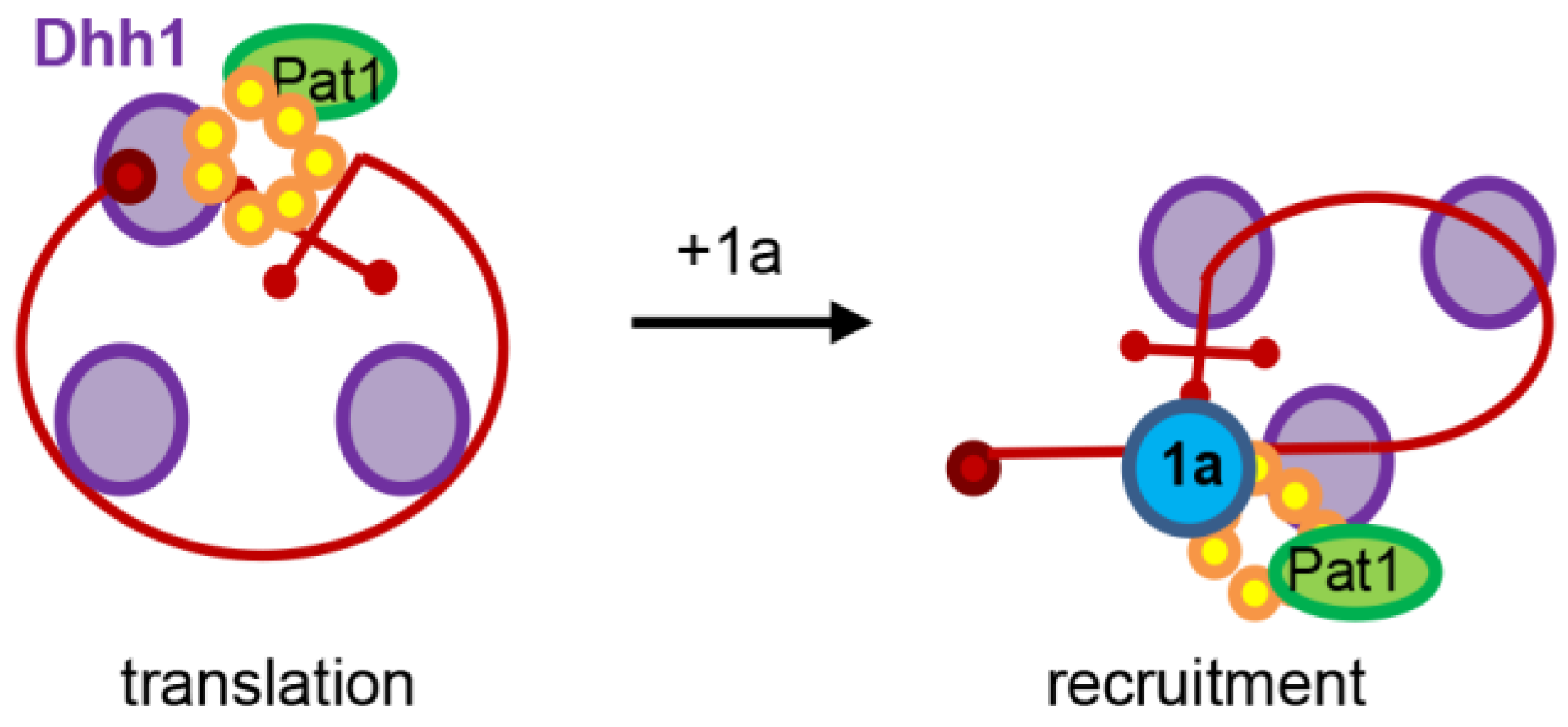
2. The Brome Mosaic Virus Converts Enemies into Collaborators in Order to Promote Viral RNA Translation and Replication
3. The Flock House Virus Subverts Features of Decapping Proteins to Control the Genomic to Sub-Genomic Viral RNA Ratio
4. Subverting Decapping Activators Is Conserved in Human (+)RNA Viruses
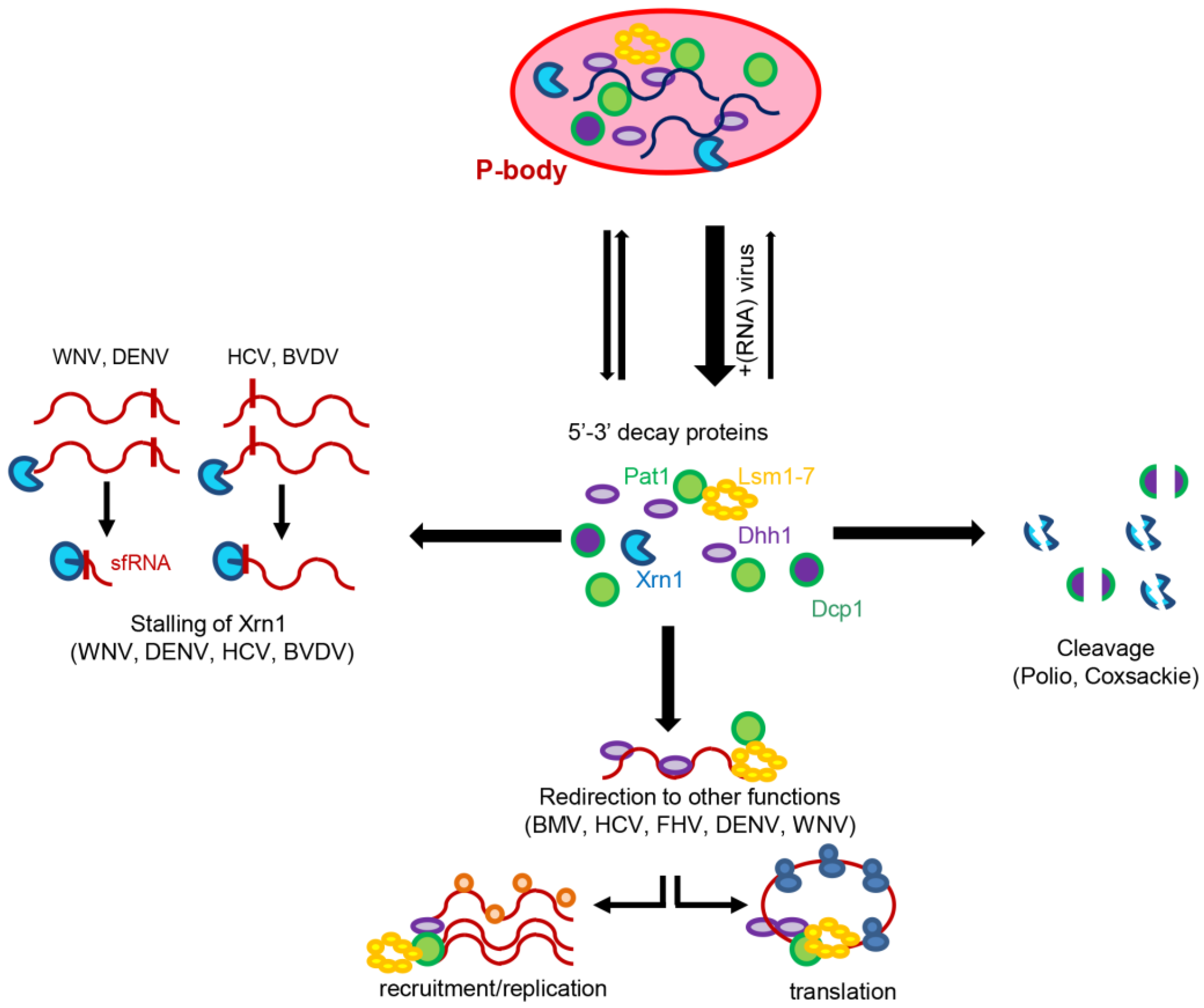
5. (+)RNA Viruses Alter the Distribution of Decay Factors
6. Concluding Remarks
Acknowledgments
Conflicts of Interest
References
- Ahlquist, P. Parallels among positive-strand RNA viruses, reverse-transcribing viruses and double-stranded RNA viruses. Nat. Rev. Microbiol. 2006, 4, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.L.; Wilusz, J. Cytoplasmic viruses: Rage against the (cellular RNA decay) machine. PLoS Pathog. 2013, 9, e1003762. [Google Scholar] [CrossRef] [PubMed]
- Parker, R. RNA degradation in saccharomyces cerevisae. Genetics 2012, 191, 671–702. [Google Scholar] [CrossRef] [PubMed]
- Daugeron, M.C.; Mauxion, F.; Seraphin, B. The yeast pop2 gene encodes a nuclease involved in mRNA deadenylation. Nucleic Acids Res. 2001, 29, 2448–2455. [Google Scholar] [CrossRef] [PubMed]
- Tucker, M.; Staples, R.R.; Valencia-Sanchez, M.A.; Muhlrad, D.; Parker, R. Ccr4p is the catalytic subunit of a ccr4p/pop2p/notp mRNA deadenylase complex in saccharomyces cerevisiae. EMBO J. 2002, 21, 1427–1436. [Google Scholar] [CrossRef] [PubMed]
- Shoemaker, C.J.; Green, R. Translation drives mRNA quality control. Nat. Struct. Mol. Biol. 2012, 19, 594–601. [Google Scholar] [CrossRef] [PubMed]
- Muhlrad, D.; Parker, R. Premature translational termination triggers mRNA decapping. Nature 1994, 370, 578–581. [Google Scholar] [CrossRef] [PubMed]
- van Hoof, A.; Frischmeyer, P.A.; Dietz, H.C.; Parker, R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science 2002, 295, 2262–2264. [Google Scholar] [CrossRef] [PubMed]
- Doma, M.K.; Parker, R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature 2006, 440, 561–564. [Google Scholar] [CrossRef] [PubMed]
- Wilusz, C.J.; Wilusz, J. Eukaryotic lsm proteins: Lessons from bacteria. Nat. Struct. Mol. Biol. 2005, 12, 1031–1036. [Google Scholar] [CrossRef] [PubMed]
- Marnef, A.; Standart, N. Pat1 proteins: A life in translation, translation repression and mRNA decay. Biochem. Soc. Trans. 2010, 38, 1602–1607. [Google Scholar] [CrossRef] [PubMed]
- Tharun, S. Purification and analysis of the decapping activator lsm1p-7p-pat1p complex from yeast. Methods Enzymol. 2008, 448, 41–55. [Google Scholar] [PubMed]
- Coller, J.; Parker, R. General translational repression by activators of mRNA decapping. Cell 2005, 122, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Nissan, T.; Rajyaguru, P.; She, M.; Song, H.; Parker, R. Decapping activators in saccharomyces cerevisiae act by multiple mechanisms. Mol. Cell 2010, 39, 773–783. [Google Scholar] [CrossRef] [PubMed]
- Rajyaguru, P.; She, M.; Parker, R. Scd6 targets eif4g to repress translation: Rgg motif proteins as a class of eif4g-binding proteins. Mol. Cell 2012, 45, 244–254. [Google Scholar] [CrossRef] [PubMed]
- Sweet, T.; Kovalak, C.; Coller, J. The dead-box protein dhh1 promotes decapping by slowing ribosome movement. PLoS Biol. 2012, 10, e1001342. [Google Scholar] [CrossRef] [PubMed]
- Aizer, A.; Kalo, A.; Kafri, P.; Shraga, A.; Ben-Yishay, R.; Jacob, A.; Kinor, N.; Shav-Tal, Y. Quantifying mRNA targeting to p-bodies in living human cells reveals their dual role in mRNA decay and storage. J. Cell Sci. 2014, 127, 4443–4456. [Google Scholar] [CrossRef] [PubMed]
- Arribere, J.A.; Doudna, J.A.; Gilbert, W.V. Reconsidering movement of eukaryotic mRNAs between polysomes and p bodies. Mol. Cell 2011, 44, 745–758. [Google Scholar] [CrossRef] [PubMed]
- Dunckley, T.; Tucker, M.; Parker, R. Two related proteins, edc1p and edc2p, stimulate mRNA decapping in saccharomyces cerevisiae. Genetics 2001, 157, 27–37. [Google Scholar] [PubMed]
- Harigaya, Y.; Jones, B.N.; Muhlrad, D.; Gross, J.D.; Parker, R. Identification and analysis of the interaction between edc3 and dcp2 in saccharomyces cerevisiae. Mol. Cell Biol. 2010, 30, 1446–1456. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, D.; Decker, C.J.; Parker, R. The enhancer of decapping proteins, edc1p and edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA 2003, 9, 239–251. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, J.D.; White, J.P.; Lloyd, R.E. Poliovirus-mediated disruption of cytoplasmic processing bodies. J. Virol. 2011, 85, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Rozovics, J.M.; Chase, A.J.; Cathcart, A.L.; Chou, W.; Gershon, P.D.; Palusa, S.; Wilusz, J.; Semler, B.L. PicoRNAvirus modification of a host mRNA decay protein. MBio 2012, 3, e00431-12. [Google Scholar] [CrossRef] [PubMed]
- Pijlman, G.P.; Funk, A.; Kondratieva, N.; Leung, J.; Torres, S.; van der Aa, L.; Liu, W.J.; Palmenberg, A.C.; Shi, P.Y.; Hall, R.A.; et al. A highly structured, nuclease-resistant, noncoding RNA produced by flaviviruses is required for pathogenicity. Cell Host Microbe 2008, 4, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Schnettler, E.; Sterken, M.G.; Leung, J.Y.; Metz, S.W.; Geertsema, C.; Goldbach, R.W.; Vlak, J.M.; Kohl, A.; Khromykh, A.A.; Pijlman, G.P. Noncoding flavivirus RNA displays RNA interference suppressor activity in insect and mammalian cells. J. Virol. 2012, 86, 13486–13500. [Google Scholar] [CrossRef] [PubMed]
- Goertz, G.P.; Fros, J.J.; Miesen, P.; Vogels, C.B.; van der Bent, M.L.; Geertsema, C.; Koenraadt, C.J.; van Rij, R.P.; van Oers, M.M.; Pijlman, G.P. Noncoding subgenomic flavivirus RNA is processed by the mosquito RNA interference machinery and determines west nile virus transmission by culex pipiens mosquitoes. J. Virol. 2016, 90, 10145–10159. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.G.; Costantino, D.A.; Rabe, J.L.; Moon, S.L.; Wilusz, J.; Nix, J.C.; Kieft, J.S. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 2014, 344, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Chapman, E.G.; Moon, S.L.; Wilusz, J.; Kieft, J.S. RNA structures that resist degradation by xrn1 produce a pathogenic dengue virus RNA. Elife 2014, 3, e01892. [Google Scholar] [CrossRef] [PubMed]
- Moon, S.L.; Anderson, J.R.; Kumagai, Y.; Wilusz, C.J.; Akira, S.; Khromykh, A.A.; Wilusz, J. A noncoding RNA produced by arthropod-borne flaviviruses inhibits the cellular exoribonuclease xrn1 and alters host mRNA stability. RNA 2012, 18, 2029–2040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moon, S.L.; Blackinton, J.G.; Anderson, J.R.; Dozier, M.K.; Dodd, B.J.; Keene, J.D.; Wilusz, C.J.; Bradrick, S.S.; Wilusz, J. Xrn1 stalling in the 5′ utr of hepatitis c virus and bovine viral diarrhea virus is associated with dysregulated host mRNA stability. PLoS Pathog. 2015, 11, e1004708. [Google Scholar] [CrossRef] [PubMed]
- Galao, R.P.; Scheller, N.; Alves-Rodrigues, I.; Breinig, T.; Meyerhans, A.; Diez, J. Saccharomyces cerevisiae: A versatile eukaryotic system in virology. Microb. Cell Fact. 2007, 6, 32. [Google Scholar] [CrossRef] [PubMed]
- Nagy, P.D. Yeast as a model host to explore plant virus-host interactions. Annu. Rev. Phytopathol. 2008, 46, 217–242. [Google Scholar] [CrossRef] [PubMed]
- Noueiry, A.O.; Ahlquist, P. Brome mosaic virus RNA replication: Revealing the role of the host in RNA virus replication. Annu. Rev. Phytopathol. 2003, 41, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Gancarz, B.L.; Hao, L.; He, Q.; Newton, M.A.; Ahlquist, P. Systematic identification of novel, essential host genes affecting bromovirus RNA replication. PLoS ONE 2011, 6, e23988. [Google Scholar] [CrossRef] [PubMed]
- Kushner, D.B.; Lindenbach, B.D.; Grdzelishvili, V.Z.; Noueiry, A.O.; Paul, S.M.; Ahlquist, P. Systematic, genome-wide identification of host genes affecting replication of a positive-strand RNA virus. Proc. Natl. Acad. Sci. USA 2003, 100, 15764–15769. [Google Scholar] [CrossRef] [PubMed]
- Alves-Rodrigues, I.; Mas, A.; Diez, J. Xenopus xp54 and human rck/p54 helicases functionally replace yeast dhh1p in brome mosaic virus RNA replication. J. Virol. 2007, 81, 4378–4380. [Google Scholar] [CrossRef] [PubMed]
- Galao, R.P.; Chari, A.; Alves-Rodrigues, I.; Lobao, D.; Mas, A.; Kambach, C.; Fischer, U.; Diez, J. Lsm1–7 complexes bind to specific sites in viral RNA genomes and regulate their translation and replication. RNA 2010, 16, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Jungfleisch, J.; Chowdhury, A.; Alves-Rodrigues, I.; Tharun, S.; Diez, J. The lsm1–7-pat1 complex promotes viral RNA translation and replication by differential mechanisms. RNA 2015, 21, 1469–1479. [Google Scholar] [CrossRef] [PubMed]
- Mas, A.; Alves-Rodrigues, I.; Noueiry, A.; Ahlquist, P.; Diez, J. Host deadenylation-dependent mRNA decapping factors are required for a key step in brome mosaic virus RNA replication. J. Virol. 2006, 80, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Diez, J.; Ishikawa, M.; Kaido, M.; Ahlquist, P. Identification and characterization of a host protein required for efficient template selection in viral RNA replication. Proc. Natl. Acad. Sci. USA 2000, 97, 3913–3918. [Google Scholar] [CrossRef] [PubMed]
- Noueiry, A.O.; Diez, J.; Falk, S.P.; Chen, J.; Ahlquist, P. Yeast lsm1p–7p/pat1p deadenylation-dependent mRNA-decapping factors are required for brome mosaic virus genomic RNA translation. Mol. Cell Biol. 2003, 23, 4094–4106. [Google Scholar] [CrossRef] [PubMed]
- Jungfleisch, J.; Nedialkova, D.D.; Dotu, I.; Sloan, K.E.; Martinez-Bosch, N.; Bruning, L.; Raineri, E.; Navarro, P.; Bohnsack, M.T.; Leidel, S.A.; et al. A novel translational control mechanism involving RNA structures within coding sequences. Genome Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Venter, P.A.; Schneemann, A. Recent insights into the biology and biomedical applications of flock house virus. Cell. Mol. Life Sci. 2008, 65, 2675–2687. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, R.; Garcia, B.H., 2nd; Goodman, R.M. Systemic spread of an RNA insect virus in plants expressing plant viral movement protein genes. Proc. Natl. Acad. Sci. USA 2001, 98, 4910–4915. [Google Scholar] [CrossRef] [PubMed]
- Johnson, K.L.; Ball, L.A. Replication of flock house virus RNAs from primary transcripts made in cells by RNA polymerase II. J. Virol. 1997, 71, 3323–3327. [Google Scholar] [PubMed]
- Lu, R.; Maduro, M.; Li, F.; Li, H.W.; Broitman-Maduro, G.; Li, W.X.; Ding, S.W. Animal virus replication and RNAi-mediated antiviral silencing in caenorhabditis elegans. Nature 2005, 436, 1040–1043. [Google Scholar] [CrossRef] [PubMed]
- Price, B.D.; Rueckert, R.R.; Ahlquist, P. Complete replication of an animal virus and maintenance of expression vectors derived from it in saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1996, 93, 9465–9470. [Google Scholar] [CrossRef] [PubMed]
- Gimenez-Barcons, M.; Alves-Rodrigues, I.; Jungfleisch, J.; Van Wynsberghe, P.M.; Ahlquist, P.; Diez, J. The cellular decapping activators lsm1, pat1, and dhh1 control the ratio of subgenomic to genomic flock house virus RNAs. J. Virol. 2013, 87, 6192–6200. [Google Scholar] [CrossRef] [PubMed]
- Eckerle, L.D.; Ball, L.A. Replication of the RNA segments of a bipartite viral genome is coordinated by a transactivating subgenomic RNA. Virology 2002, 296, 165–176. [Google Scholar] [CrossRef] [PubMed]
- Lindenbach, B.D.; Sgro, J.Y.; Ahlquist, P. Long-distance base pairing in flock house virus RNA1 regulates subgenomic RNA3 synthesis and RNA2 replication. J. Virol. 2002, 76, 3905–3919. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Rueckert, R.R. Flock house virus: Down-regulation of subgenomic RNA3 synthesis does not involve coat protein and is targeted to synthesis of its positive strand. J. Virol. 1993, 67, 2716–2722. [Google Scholar] [PubMed]
- Weston, A.; Sommerville, J. Xp54 and related (ddx6-like) RNA helicases: Roles in messenger rnp assembly, translation regulation and RNA degradation. Nucl. Acids Res. 2006, 34, 3082–3094. [Google Scholar] [CrossRef] [PubMed]
- Scheller, N.; Mina, L.B.; Galao, R.P.; Chari, A.; Gimenez-Barcons, M.; Noueiry, A.; Fischer, U.; Meyerhans, A.; Diez, J. Translation and replication of hepatitis c virus genomic RNA depends on ancient cellular proteins that control mRNA fates. Proc. Natl. Acad. Sci. USA 2009, 106, 13517–13522. [Google Scholar] [CrossRef] [PubMed]
- Huys, A.; Thibault, P.A.; Wilson, J.A. Modulation of hepatitis c virus RNA accumulation and translation by ddx6 and mir-122 are mediated by separate mechanisms. PLoS ONE 2013, 8, e67437. [Google Scholar] [CrossRef] [PubMed]
- Jangra, R.K.; Yi, M.; Lemon, S.M. Ddx6 (rck/p54) is required for efficient hepatitis c virus replication but not for internal ribosome entry site-directed translation. J. Virol. 2010, 84, 6810–6824. [Google Scholar] [CrossRef] [PubMed]
- Chahar, H.S.; Chen, S.; Manjunath, N. P-body components lsm1, gw182, ddx3, ddx6 and xrn1 are recruited to wnv replication sites and positively regulate viral replication. Virology 2013, 436, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Ward, A.M.; Bidet, K.; Yinglin, A.; Ler, S.G.; Hogue, K.; Blackstock, W.; Gunaratne, J.; Garcia-Blanco, M.A. Quantitative mass spectrometry of denv-2 RNA-interacting proteins reveals that the dead-box RNA helicase ddx6 binds the db1 and db2 3′ utr structures. RNA Biol. 2011, 8, 1173–1186. [Google Scholar] [CrossRef] [PubMed]
- Schuppli, D.; Miranda, G.; Tsui, H.C.; Winkler, M.E.; Sogo, J.M.; Weber, H. Altered 3′-terminal RNA structure in phage qbeta adapted to host factor-less escherichia coli. Proc. Natl. Acad. Sci. USA 1997, 94, 10239–10242. [Google Scholar] [PubMed]
- Buchan, J.R. Mrnp granules: Assembly, function, and connections with disease. RNA Biol. 2014, 11. [Google Scholar] [CrossRef] [PubMed]
- Jain, S.; Parker, R. The discovery and analysis of p bodies. Adv. Exp. Med. Biol. 2013, 768, 23–43. [Google Scholar] [PubMed]
- Cougot, N.; Babajko, S.; Seraphin, B. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 2004, 165, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Brengues, M.; Teixeira, D.; Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 2005, 310, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Andrei, M.A.; Ingelfinger, D.; Heintzmann, R.; Achsel, T.; Rivera-Pomar, R.; Luhrmann, R. A role for eif4e and eif4e-transporter in targeting mrnps to mammalian processing bodies. RNA 2005, 11, 717–727. [Google Scholar] [CrossRef] [PubMed]
- Aizer, A.; Brody, Y.; Ler, L.W.; Sonenberg, N.; Singer, R.H.; Shav-Tal, Y. The dynamics of mammalian p body transport, assembly, and disassembly in vivo. Mol. Biol. Cell 2008, 19, 4154–4166. [Google Scholar] [CrossRef] [PubMed]
- Kedersha, N.; Stoecklin, G.; Ayodele, M.; Yacono, P.; Lykke-Andersen, J.; Fritzler, M.J.; Scheuner, D.; Kaufman, R.J.; Golan, D.E.; Anderson, P. Stress granules and processing bodies are dynamically linked sites of mrnp remodeling. J. Cell Biol. 2005, 169, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.H.; Yu, J.H.; Gulick, T.; Bloch, K.D.; Bloch, D.B. RNA-associated protein 55 (rap55) localizes to mRNA processing bodies and stress granules. RNA 2006, 12, 547–554. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.H.; Yang, W.H.; Gulick, T.; Bloch, K.D.; Bloch, D.B. Ge-1 is a central component of the mammalian cytoplasmic mRNA processing body. RNA 2005, 11, 1795–1802. [Google Scholar] [CrossRef] [PubMed]
- Ozgur, S.; Chekulaeva, M.; Stoecklin, G. Human pat1b connects deadenylation with mRNA decapping and controls the assembly of processing bodies. Mol. Cell Biol. 2010, 30, 4308–4323. [Google Scholar] [CrossRef] [PubMed]
- Ayache, J.; Benard, M.; Ernoult-Lange, M.; Minshall, N.; Standart, N.; Kress, M.; Weil, D. P-body assembly requires ddx6 repression complexes rather than decay or ataxin2/2l complexes. Mol. Biol. Cell 2015, 26, 2579–2595. [Google Scholar] [CrossRef] [PubMed]
- Perez-Vilaro, G.; Scheller, N.; Saludes, V.; Diez, J. Hepatitis c virus infection alters p-body composition but is independent of p-body granules. J. Virol. 2012, 86, 8740–8749. [Google Scholar] [CrossRef] [PubMed]
- Emara, M.M.; Brinton, M.A. Interaction of tia-1/tiar with west nile and dengue virus products in infected cells interferes with stress granule formation and processing body assembly. Proc. Natl. Acad. Sci. USA 2007, 104, 9041–9046. [Google Scholar] [CrossRef] [PubMed]
- Ariumi, Y.; Kuroki, M.; Kushima, Y.; Osugi, K.; Hijikata, M.; Maki, M.; Ikeda, M.; Kato, N. Hepatitis c virus hijacks p-body and stress granule components around lipid droplets. J. Virol. 2011, 85, 6882–6892. [Google Scholar] [CrossRef] [PubMed]
- Perez-Vilaro, G.; Fernandez-Carrillo, C.; Mensa, L.; Miquel, R.; Sanjuan, X.; Forns, X.; Perez-del-Pulgar, S.; Diez, J. Hepatitis c virus infection inhibits p-body granule formation in human livers. J. Hepatol. 2015, 62, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Eulalio, A.; Behm-Ansmant, I.; Schweizer, D.; Izaurralde, E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell Biol. 2007, 27, 3970–3981. [Google Scholar] [CrossRef] [PubMed]

© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jungfleisch, J.; Blasco-Moreno, B.; Díez, J. Use of Cellular Decapping Activators by Positive-Strand RNA Viruses. Viruses 2016, 8, 340. https://doi.org/10.3390/v8120340
Jungfleisch J, Blasco-Moreno B, Díez J. Use of Cellular Decapping Activators by Positive-Strand RNA Viruses. Viruses. 2016; 8(12):340. https://doi.org/10.3390/v8120340
Chicago/Turabian StyleJungfleisch, Jennifer, Bernat Blasco-Moreno, and Juana Díez. 2016. "Use of Cellular Decapping Activators by Positive-Strand RNA Viruses" Viruses 8, no. 12: 340. https://doi.org/10.3390/v8120340




