Determinants of Genomic RNA Encapsidation in the Saccharomyces cerevisiae Long Terminal Repeat Retrotransposons Ty1 and Ty3
Abstract
:1. Introduction
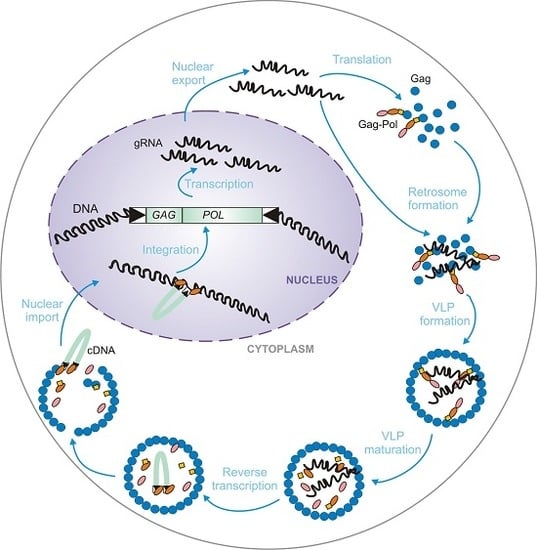
2. Genetic Organization and Replication Cycle of Ty Retrotransposons
3. Cis-Acting Sequences in Retrotransposons gRNA
4. Retrotransposon Gag Proteins
5. Trafficking of Ty gRNA and Gag to Retrosomes
6. P-Bodies and Retrosome Formation
7. RNA Packaging
8. Gag Assembly into VLPs
9. Gag/RNA Interactions Important for Copy Number Control (CNC)
10. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Havecker, E.R.; Gao, X.; Voytas, D.F. The diversity of LTR retrotransposons. Genome Biol. 2004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jern, P.; Coffin, J.M. Effects of retroviruses on host genome function. Annu. Rev. Genet. 2008, 42, 709–732. [Google Scholar] [CrossRef] [PubMed]
- Llorens, C.; Fares, M.A.; Moya, A. Relationships of gag-pol diversity between Ty3/Gypsy and retroviridae LTR retroelements and the three kings hypothesis. BMC Evol. Biol. 2008. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.S.; Henikoff, S.; Eickbush, T.H. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res. 2000, 10, 1307–1318. [Google Scholar] [CrossRef] [PubMed]
- Voytas, D.F.; Boeke, J.D. Ty1 and Ty5 of Sacharomyces cerevisiae. In Mobile DNA II; Craig, N.L., Craigie, R., Gellert, M., Lambowitz, A.M., Eds.; ASM Press: Washington, DC, USA, 2002; pp. 614–630. [Google Scholar]
- Boeke, J.D.; Garfinkel, D.J.; Styles, C.A.; Fink, G.R. Ty elements transpose through an RNA intermediate. Cell 1985, 40, 491–500. [Google Scholar] [CrossRef]
- Clark, D.J.; Bilanchone, V.W.; Haywood, L.J.; Dildine, S.L.; Sandmeyer, S.B. A yeast sigma composite element, Ty3, has properties of a retrotransposon. J. Biol. Chem. 1988, 263, 1413–1423. [Google Scholar] [PubMed]
- Garfinkel, D.J.; Boeke, J.D.; Fink, G.R. Ty element transposition: Reverse transcriptase and virus-like particles. Cell 1985, 42, 507–517. [Google Scholar] [CrossRef]
- Garfinkel, D.J. Genome evolution mediated by ty elements in Saccharomyces. Cytogenet. Genome Res. 2005, 110, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Mita, P.; Boeke, J.D. How retrotransposons shape genome regulation. Curr. Opin. Genet. Dev. 2016, 37, 90–100. [Google Scholar] [CrossRef] [PubMed]
- Curcio, M.J.; Lutz, S.; Lesage, P. The Ty1 LTR-retrotransposon of budding yeast. Microbiol. Spectr. 2015, 3, 1–35. [Google Scholar] [CrossRef] [PubMed]
- Sandmeyer, S.; Patterson, K.; Bilanchone, V. Ty3, a position-specific retrotransposon in budding yeast. Microbiol. Spectr. 2015. [Google Scholar] [CrossRef] [PubMed]
- Curcio, M.J.; Garfinkel, D.J. Heterogeneous functional Ty1 elements are abundant in the saccharomyces cerevisiae genome. Genetics 1994, 136, 1245–1259. [Google Scholar] [PubMed]
- Elder, R.T.; St John, T.P.; Stinchcomb, D.T.; Davis, R.W.; Scherer, S.; Davis, R.W. Studies on the transposable element Ty1 of yeast. I. RNA homologous to Ty1. II. Recombination and expression of Ty1 and adjacent sequences. Cold Spring Harb. Symp. Quant. Biol. 1981, 45, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Carr, M.; Bensasson, D.; Bergman, C.M. Evolutionary genomics of transposable elements in saccharomyces cerevisiae. PLoS ONE 2012, 7, e50978. [Google Scholar] [CrossRef] [PubMed]
- Bilanchone, V.W.; Claypool, J.A.; Kinsey, P.T.; Sandmeyer, S.B. Positive and negative regulatory elements control expression of the yeast retrotransposon Ty3. Genetics 1993, 134, 685–700. [Google Scholar] [PubMed]
- Hansen, L.J.; Chalker, D.L.; Sandmeyer, S.B. Ty3, a yeast retrotransposon associated with tRNA genes, has homology to animal retroviruses. Mol. Cell. Biol. 1988, 8, 5245–5256. [Google Scholar] [CrossRef] [PubMed]
- Menees, T.M.; Sandmeyer, S.B. Transposition of the yeast retroviruslike element Ty3 is dependent on the cell cycle. Mol. Cell. Biol. 1994, 14, 8229–8240. [Google Scholar] [CrossRef] [PubMed]
- Mellor, J.; Fulton, A.M.; Dobson, M.J.; Roberts, N.A.; Wilson, W.; Kingsman, A.J.; Kingsman, S.M. The Ty transposon of saccharomyces cerevisiae determines the synthesis of at least three proteins. Nucleic Acids Res. 1985, 13, 6249–6263. [Google Scholar] [CrossRef] [PubMed]
- Elder, R.T.; Loh, E.Y.; Davis, R.W. RNA from the yeast transposable element Ty1 has both ends in the direct repeats, a structure similar to retrovirus RNA. Proc. Natl. Acad. Sci. USA 1983, 80, 2432–2436. [Google Scholar] [CrossRef] [PubMed]
- Mules, E.H.; Uzun, O.; Gabriel, A. In vivo Ty1 reverse transcription can generate replication intermediates with untidy ends. J. Virol. 1998, 72, 6490–6503. [Google Scholar] [PubMed]
- Eickbush, T.H.; Jamburuthugoda, V.K. The diversity of retrotransposons and the properties of their reverse transcriptases. Virus Res. 2008, 134, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Hansen, L.J.; Chalker, D.L.; Orlinsky, K.J.; Sandmeyer, S.B. Ty3 Gag3 and POL3 genes encode the components of intracellular particles. J. Virol. 1992, 66, 1414–1424. [Google Scholar] [PubMed]
- Hansen, L.J.; Sandmeyer, S.B. Characterization of a transpositionally active Ty3 element and identification of the Ty3 integrase protein. J. Virol. 1990, 64, 2599–2607. [Google Scholar] [PubMed]
- Curcio, M.J.; Hedge, A.M.; Boeke, J.D.; Garfinkel, D.J. Ty RNA levels determine the spectrum of retrotransposition events that activate gene expression in saccharomyces cerevisiae. Mol. Gen. Genet. 1990, 220, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Curcio, M.J.; Sanders, N.J.; Garfinkel, D.J. Transpositional competence and transcription of endogenous ty elements in Saccharomyces cerevisiae: Implications for regulation of transposition. Mol. Cell. Biol. 1988, 8, 3571–3581. [Google Scholar] [CrossRef] [PubMed]
- Munchel, S.E.; Shultzaberger, R.K.; Takizawa, N.; Weis, K. Dynamic profiling of mRNA turnover reveals gene-specific and system-wide regulation of mRNA decay. Mol. Biol. Cell 2011, 22, 2787–2795. [Google Scholar] [CrossRef] [PubMed]
- Checkley, M.A.; Mitchell, J.A.; Eizenstat, L.D.; Lockett, S.J.; Garfinkel, D.J. Ty1 gag enhances the stability and nuclear export of Ty1 mRNA. Traffic 2013, 14, 57–69. [Google Scholar] [CrossRef] [PubMed]
- Berretta, J.; Pinskaya, M.; Morillon, A. A cryptic unstable transcript mediates transcriptional trans-silencing of the Ty1 retrotransposon in S. cerevisiae. Genes Dev. 2008, 22, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Mitchell, J.A.; Nishida, Y.; Hildreth, J.E.; Ariberre, J.A.; Gilbert, W.V.; Garfinkel, D.J. A trans-dominant form of gag restricts Ty1 retrotransposition and mediates copy number control. J. Virol. 2015, 89, 3922–3938. [Google Scholar] [CrossRef] [PubMed]
- Winston, F.; Durbin, K.J.; Fink, G.R. The SPT3 gene is required for normal transcription of Ty elements in S. cerevisiae. Cell 1984, 39, 675–682. [Google Scholar] [CrossRef]
- Yu, K.; Elder, R.T. Some of the signals for 3′-end formation in transcription of the Saccharomyces cerevisiae Ty-d15 element are immediately downstream of the initiation site. Mol. Cell. Biol. 1989, 9, 2431–2444. [Google Scholar] [CrossRef] [PubMed]
- Belcourt, M.F.; Farabaugh, P.J. Ribosomal frameshifting in the yeast retrotransposon Ty: TRNAs induce slippage on a 7 nucleotide minimal site. Cell 1990, 62, 339–352. [Google Scholar] [CrossRef]
- Clare, J.J.; Belcourt, M.; Farabaugh, P.J. Efficient translational frameshifting occurs within a conserved sequence of the overlap between the two genes of a yeast Ty1 transposon. Proc. Natl. Acad. Sci. USA 1988, 85, 6816–6820. [Google Scholar] [CrossRef] [PubMed]
- Farabaugh, P.J.; Zhao, H.; Vimaladithan, A. A novel programed frameshift expresses the POL3 gene of retrotransposon Ty3 of yeast: Frameshifting without tRNA slippage. Cell 1993, 74, 93–103. [Google Scholar] [CrossRef]
- Kirchner, J.; Sandmeyer, S.B.; Forrest, D.B. Transposition of a Ty3 GAG3-POL3 fusion mutant is limited by availability of capsid protein. J. Virol. 1992, 66, 6081–6092. [Google Scholar] [PubMed]
- Vimaladithan, A.; Farabaugh, P.J. Special peptidyl-tRNA molecules can promote translational frameshifting without slippage. Mol. Cell. Biol. 1994, 14, 8107–8116. [Google Scholar] [CrossRef] [PubMed]
- Kawakami, K.; Pande, S.; Faiola, B.; Moore, D.P.; Boeke, J.D.; Farabaugh, P.J.; Strathern, J.N.; Nakamura, Y.; Garfinkel, D.J. A rare tRNA-Arg(CCU) that regulates Ty1 element ribosomal frameshifting is essential for Ty1 retrotransposition in Saccharomyces cerevisiae. Genetics 1993, 135, 309–320. [Google Scholar] [PubMed]
- Xu, H.; Boeke, J.D. Host genes that influence transposition in yeast: The abundance of a rare tRNA regulates Ty1 transposition frequency. Proc. Natl. Acad. Sci. USA 1990, 87, 8360–8364. [Google Scholar] [CrossRef] [PubMed]
- Beliakova-Bethell, N.; Beckham, C.; Giddings, T.H., Jr.; Winey, M.; Parker, R.; Sandmeyer, S. Virus-like particles of the Ty3 retrotransposon assemble in association with P-body components. RNA 2006, 12, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Malagon, F.; Jensen, T.H. The T body, a new cytoplasmic RNA granule in Saccharomyces cerevisiae. Mol. Cell. Biol. 2008, 28, 6022–6032. [Google Scholar] [CrossRef] [PubMed]
- Malagon, F.; Jensen, T.H. T-body formation precedes virus-like particle maturation in S. cerevisiae. RNA Biol. 2011, 8, 184–189. [Google Scholar] [CrossRef] [PubMed]
- Sandmeyer, S.B.; Clemens, K.A. Function of a retrotransposon nucleocapsid protein. RNA Biol. 2010, 7, 642–654. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.X.; Moore, S.P.; Garfinkel, D.J.; Rein, A. The genomic RNA in Ty1 virus-like particles is dimeric. J. Virol. 2000, 74, 10819–10821. [Google Scholar] [CrossRef] [PubMed]
- Nymark-McMahon, M.H.; Beliakova-Bethell, N.S.; Darlix, J.L.; le Grice, S.F.; Sandmeyer, S.B. Ty3 integrase is required for initiation of reverse transcription. J. Virol. 2002, 76, 2804–2816. [Google Scholar] [CrossRef] [PubMed]
- Chapman, K.B.; Bystrom, A.S.; Boeke, J.D. Initiator methionine tRNA is essential for Ty1 transposition in yeast. Proc. Natl. Acad. Sci. USA 1992, 89, 3236–3240. [Google Scholar] [CrossRef] [PubMed]
- Gabus, C.; Ficheux, D.; Rau, M.; Keith, G.; Sandmeyer, S.; Darlix, J.L. The yeast Ty3 retrotransposon contains a 5′-3′ bipartite primer-binding site and encodes nucleocapsid protein NCp9 functionally homologous to HIV-1 NCp7. EMBO J. 1998, 17, 4873–4880. [Google Scholar] [CrossRef] [PubMed]
- Keeney, J.B.; Chapman, K.B.; Lauermann, V.; Voytas, D.F.; Aström, S.U.; von Pawel-Rammingen, U.; Bystrom, A.; Boeke, J.D. Multiple molecular determinants for retrotransposition in a primer tRNA. Mol. Cell. Biol. 1995, 15, 217–226. [Google Scholar] [CrossRef] [PubMed]
- Kenna, M.A.; Brachmann, C.B.; Devine, S.E.; Boeke, J.D. Invading the yeast nucleus: A nuclear localization signal at the C terminus of Ty1 integrase is required for transposition in vivo. Mol. Cell. Biol. 1998, 18, 1115–1124. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.S.; Nymark-McMahon, M.H.; Yieh, L.; Sandmeyer, S.B. Integrase mediates nuclear localization of Ty3. Mol. Cell. Biol. 2001, 21, 7826–7838. [Google Scholar] [CrossRef] [PubMed]
- Devine, S.E.; Boeke, J.D. Integration of the yeast retrotransposon Ty1 is targeted to regions upstream of genes transcribed by RNA polymerase III. Genes Dev. 1996, 10, 620–633. [Google Scholar] [CrossRef] [PubMed]
- Eigel, A.; Feldmann, H. Ty1 and delta elements occur adjacent to several tRNA genes in yeast. EMBO J. 1982, 1, 1245–1250. [Google Scholar] [PubMed]
- Kim, J.M.; Vanguri, S.; Boeke, J.D.; Gabriel, A.; Voytas, D.F. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete saccharomyces cerevisiae genome sequence. Genome Res. 1998, 8, 464–478. [Google Scholar] [PubMed]
- Qi, X.; Daily, K.; Nguyen, K.; Wang, H.; Mayhew, D.; Rigor, P.; Forouzan, S.; Johnston, M.; Mitra, R.D.; Baldi, P.; et al. Retrotransposon profiling of RNA polymerase III initiation sites. Genome Res. 2012, 22, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Connolly, C.M.; Sandmeyer, S.B. RNA polymerase III interferes with Ty3 integration. FEBS Lett. 1997, 405, 305–311. [Google Scholar] [CrossRef]
- Kirchner, J.; Connolly, C.M.; Sandmeyer, S.B. Requirement of RNA polymerase iii transcription factors for in vitro position-specific integration of a retroviruslike element. Science 1995, 267, 1488–1491. [Google Scholar] [CrossRef] [PubMed]
- Yieh, L.; Hatzis, H.; Kassavetis, G.; Sandmeyer, S.B. Mutational analysis of the transcription factor IIIB-DNA target of Ty3 retroelement integration. J. Biol. Chem. 2002, 277, 25920–25928. [Google Scholar] [CrossRef] [PubMed]
- Yieh, L.; Kassavetis, G.; Geiduschek, E.P.; Sandmeyer, S.B. The Brf and TATA-binding protein subunits of the RNA polymerase III transcription factor IIIB mediate position-specific integration of the gypsy-like element, Ty3. J. Biol. Chem. 2000, 275, 29800–29807. [Google Scholar] [CrossRef] [PubMed]
- Bridier-Nahmias, A.; Tchalikian-Cosson, A.; Baller, J.A.; Menouni, R.; Fayol, H.; Flores, A.; Saib, A.; Werner, M.; Voytas, D.F.; Lesage, P. Retrotransposons. An RNA polymerase III subunit determines sites of retrotransposon integration. Science 2015, 348, 585–588. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Boeke, J.D. Localization of sequences required in cis for yeast Ty1 element transposition near the long terminal repeats: Analysis of mini-Ty1 elements. Mol. Cell. Biol. 1990, 10, 2695–2702. [Google Scholar] [CrossRef] [PubMed]
- Purzycka, K.J.; Legiewicz, M.; Matsuda, E.; Eizentstat, L.D.; Lusvarghi, S.; Saha, A.; le Grice, S.F.; Garfinkel, D.J. Exploring Ty1 retrotransposon RNA structure within virus-like particles. Nucleic Acids Res. 2013, 41, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Nishida, Y.; Pachulska-Wieczorek, K.; Blaszczyk, L.; Saha, A.; Gumna, J.; Garfinkel, D.J.; Purzycka, K.J. Ty1 retrovirus-like element Gag contains overlapping restriction factor and nucleic acid chaperone functions. Nucleic Acids Res. 2015, 43, 7414–7431. [Google Scholar] [CrossRef] [PubMed]
- Cristofari, G.; Bampi, C.; Wilhelm, M.; Wilhelm, F.X.; Darlix, J.L. A 5′-3′ long-range interaction in Ty1 RNA controls its reverse transcription and retrotransposition. EMBO J. 2002, 21, 4368–4379. [Google Scholar] [CrossRef] [PubMed]
- Friant, S.; Heyman, T.; Wilhelm, M.L.; Wilhelm, F.X. Extended interactions between the primer tRNAi(Met) and genomic RNA of the yeast Ty1 retrotransposon. Nucleic Acids Res. 1996, 24, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, M.; Wilhelm, F.X.; Keith, G.; Agoutin, B.; Heyman, T. Yeast Ty1 retrotransposon: The minus-strand primer binding site and a cis-acting domain of the Ty1 RNA are both important for packaging of primer tRNA inside virus-like particles. Nucleic Acids Res. 1994, 22, 4560–4565. [Google Scholar] [CrossRef] [PubMed]
- Friant, S.; Heyman, T.; Bystrom, A.S.; Wilhelm, M.; Wilhelm, F.X. Interactions between Ty1 retrotransposon RNA and the T and D regions of the tRNA(iMet) primer are required for initiation of reverse transcription in vivo. Mol. Cell. Biol. 1998, 18, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Bolton, E.C.; Coombes, C.; Eby, Y.; Cardell, M.; Boeke, J.D. Identification and characterization of critical cis-acting sequences within the yeast Ty1 retrotransposon. RNA 2005, 11, 308–322. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Purzycka, K.J.; Lusvarghi, S.; Li, D.; Legrice, S.F.; Boeke, J.D. Retrotransposon Ty1 RNA contains a 5′-terminal long-range pseudoknot required for efficient reverse transcription. RNA 2013, 19, 320–332. [Google Scholar] [CrossRef] [PubMed]
- Purzycka, K.J.; Pachulska-Wieczorek, K.; Adamiak, R.W. The in vitro loose dimer structure and rearrangements of the HIV-2 leader RNA. Nucleic Acids Res. 2011, 39, 7234–7248. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.M.; Dang, K.K.; Gorelick, R.J.; Leonard, C.W.; Bess, J.W., Jr.; Swanstrom, R.; Burch, C.L.; Weeks, K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009, 460, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Kertesz, M.; Wan, Y.; Mazor, E.; Rinn, J.L.; Nutter, R.C.; Chang, H.Y.; Segal, E. Genome-wide measurement of RNA secondary structure in yeast. Nature 2010, 467, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Larsen, L.S.; Beliakova-Bethell, N.; Bilanchone, V.; Zhang, M.; Lamsa, A.; Dasilva, R.; Hatfield, G.W.; Nagashima, K.; Sandmeyer, S. Ty3 nucleocapsid controls localization of particle assembly. J. Virol. 2008, 82, 2501–2514. [Google Scholar] [CrossRef] [PubMed]
- Larsen, L.S.; Kuznetsov, Y.; McPherson, A.; Hatfield, G.W.; Sandmeyer, S. TY3 GAG3 protein forms ordered particles in Escherichia coli. Virology 2008, 370, 223–227. [Google Scholar] [CrossRef] [PubMed]
- Larsen, L.S.; Zhang, M.; Beliakova-Bethell, N.; Bilanchone, V.; Lamsa, A.; Nagashima, K.; Najdi, R.; Kosaka, K.; Kovacevic, V.; Cheng, J.; et al. Ty3 capsid mutations reveal early and late functions of the amino-terminal domain. J. Virol. 2007, 81, 6957–6972. [Google Scholar] [CrossRef] [PubMed]
- Clemens, K.; Larsen, L.; Zhang, M.; Kuznetsov, Y.; Bilanchone, V.; Randall, A.; Harned, A.; Dasilva, R.; Nagashima, K.; McPherson, A.; et al. The Ty3 Gag3 spacer controls intracellular condensation and uncoating. J. Virol. 2011, 85, 3055–3066. [Google Scholar] [CrossRef] [PubMed]
- Orlinsky, K.J.; Sandmeyer, S.B. The Cys-His motif of Ty3 NC can be contributed by Gag3 or Gag3-Pol3 polyproteins. J. Virol. 1994, 68, 4152–4166. [Google Scholar] [PubMed]
- Mirambeau, G.; Lyonnais, S.; Gorelick, R.J. Features, processing states, and heterologous protein interactions in the modulation of the retroviral nucleocapsid protein function. RNA Biol. 2010, 7, 724–734. [Google Scholar] [CrossRef] [PubMed]
- Chaurasiya, K.R.; Geertsema, H.; Cristofari, G.; Darlix, J.L.; Williams, M.C. A single zinc finger optimizes the DNA interactions of the nucleocapsid protein of the yeast retrotransposon Ty3. Nucleic Acids Res. 2012, 40, 751–760. [Google Scholar] [CrossRef] [PubMed]
- Rein, A. Nucleic acid chaperone activity of retroviral Gag proteins. RNA Biol. 2010, 7, 700–705. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.X.; Campbell, S.; Harvin, D.; Ehresmann, B.; Ehresmann, C.; Rein, A. The human immunodeficiency virus Type 1 Gag polyprotein has nucleic acid chaperone activity: Possible role in dimerization of genomic RNA and placement of tRNA on the primer binding site. J. Virol. 1999, 73, 4251–4256. [Google Scholar] [PubMed]
- Pachulska-Wieczorek, K.; Blaszczyk, L.; Biesiada, M.; Adamiak, R.W.; Purzycka, K.J. The matrix domain contributes to the nucleic acid chaperone activity of HIV-2 Gag. Retrovirology 2016. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Datta, S.A.; Mitra, M.; Gorelick, R.J.; Rein, A.; Levin, J.G. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: Biological implications. Virology 2010, 405, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.E.; Mellor, J.; Gull, K.; Sim, R.B.; Tuite, M.F.; Kingsman, S.M.; Kingsman, A.J. The functions and relationships of Ty-VLP proteins in yeast reflect those of mammalian retroviral proteins. Cell 1987, 49, 111–119. [Google Scholar] [CrossRef]
- Garfinkel, D.J.; Hedge, A.M.; Youngren, S.D.; Copeland, T.D. Proteolytic processing of pol-TYB proteins from the yeast retrotransposon Ty1. J. Virol. 1991, 65, 4573–4581. [Google Scholar] [PubMed]
- HA, A.L.-K.; Bhella, D.; Kenney, J.M.; Roth, J.F.; Kingsman, A.J.; Martin-Rendon, E.; Saibil, H.R. Yeast Ty retrotransposons assemble into virus-like particles whose T-numbers depend on the C-terminal length of the capsid protein. J. Mol. Biol. 1999, 292, 65–73. [Google Scholar]
- Merkulov, G.V.; Swiderek, K.M.; Brachmann, C.B.; Boeke, J.D. A critical proteolytic cleavage site near the C terminus of the yeast retrotransposon Ty1 Gag protein. J. Virol. 1996, 70, 5548–5556. [Google Scholar] [PubMed]
- Youngren, S.D.; Boeke, J.D.; Sanders, N.J.; Garfinkel, D.J. Functional organization of the retrotransposon Ty from saccharomyces cerevisiae: Ty protease is required for transposition. Mol. Cell. Biol. 1988, 8, 1421–1431. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.F.; Kingsman, S.M.; Kingsman, A.J.; Martin-Rendon, E. Possible regulatory function of the Saccharomyces cerevisiae Ty1 retrotransposon core protein. Yeast 2000, 16, 921–932. [Google Scholar] [CrossRef]
- Cristofari, G.; Ficheux, D.; Darlix, J.L. The Gag-like protein of the yeast Ty1 retrotransposon contains a nucleic acid chaperone domain analogous to retroviral nucleocapsid proteins. J. Biol. Chem. 2000, 275, 19210–19217. [Google Scholar] [CrossRef] [PubMed]
- De Rocquigny, H.; Gabus, C.; Vincent, A.; Fournie-Zaluski, M.C.; Roques, B.; Darlix, J.L. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl. Acad. Sci. USA 1992, 89, 6472–6476. [Google Scholar] [CrossRef] [PubMed]
- Housset, V.; De Rocquigny, H.; Roques, B.P.; Darlix, J.L. Basic amino acids flanking the zinc finger of moloney murine leukemia virus nucleocapsid protein NCp10 are critical for virus infectivity. J. Virol. 1993, 67, 2537–2545. [Google Scholar] [PubMed]
- Le Cam, E.; Coulaud, D.; Delain, E.; Petitjean, P.; Roques, B.P.; Gerard, D.; Stoylova, E.; Vuilleumier, C.; Stoylov, S.P.; Mely, Y. Properties and growth mechanism of the ordered aggregation of a model RNA by the HIV-1 nucleocapsid protein: An electron microscopy investigation. Biopolymers 1998, 45, 217–229. [Google Scholar] [CrossRef]
- Pachulska-Wieczorek, K.; Stefaniak, A.K.; Purzycka, K.J. Similarities and differences in the nucleic acid chaperone activity of HIV-2 and HIV-1 nucleocapsid proteins in vitro. Retrovirology 2014. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Mitra, M.; Naufer, M.N.; McCauley, M.J.; Gorelick, R.J.; Rouzina, I.; Musier-Forsyth, K.; Williams, M.C. Differential contribution of basic residues to HIV-1 nucleocapsid protein’s nucleic acid chaperone function and retroviral replication. Nucleic Acids Res. 2014, 42, 2525–2537. [Google Scholar] [CrossRef] [PubMed]
- Doh, J.H.; Lutz, S.; Curcio, M.J. Co-translational localization of an LTR-retrotransposon RNA to the endoplasmic reticulum nucleates virus-like particle assembly sites. PLoS Genet. 2014, 10, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Hermesh, O.; Jansen, R.P. Take the (RN)a-train: Localization of mRNA to the endoplasmic reticulum. Biochim. Biophys. Acta 2013, 1833, 2519–2525. [Google Scholar] [CrossRef] [PubMed]
- Checkley, M.A.; Nagashima, K.; Lockett, S.J.; Nyswaner, K.M.; Garfinkel, D.J. P-body components are required for Ty1 retrotransposition during assembly of retrotransposition-competent virus-like particles. Mol. Cell. Biol. 2010, 30, 382–398. [Google Scholar] [CrossRef] [PubMed]
- Dupont, S.; Sharova, N.; DeHoratius, C.; Virbasius, C.M.; Zhu, X.; Bukrinskaya, A.G.; Stevenson, M.; Green, M.R. A novel nuclear export activity in HIV-1 matrix protein required for viral replication. Nature 1999, 402, 681–685. [Google Scholar] [PubMed]
- Nash, M.A.; Meyer, M.K.; Decker, G.L.; Arlinghaus, R.B. A subset of Pr65gag is nucleus associated in murine leukemia virus-infected cells. J. Virol. 1993, 67, 1350–1356. [Google Scholar] [PubMed]
- Schliephake, A.W.; Rethwilm, A. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 1994, 68, 4946–4954. [Google Scholar] [PubMed]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 3944–3949. [Google Scholar] [CrossRef] [PubMed]
- Garbitt-Hirst, R.; Kenney, S.P.; Parent, L.J. Genetic evidence for a connection between Rous sarcoma virus Gag nuclear trafficking and genomic RNA packaging. J. Virol. 2009, 83, 6790–6797. [Google Scholar] [CrossRef] [PubMed]
- Aizer, A.; Brody, Y.; Ler, L.W.; Sonenberg, N.; Singer, R.H.; Shav-Tal, Y. The dynamics of mammalian P body transport, assembly, and disassembly in vivo. Mol. Biol. Cell 2008, 19, 4154–4166. [Google Scholar] [CrossRef] [PubMed]
- Anderson, P.; Kedersha, N. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell. Biol. 2009, 10, 430–436. [Google Scholar] [CrossRef] [PubMed]
- Buchan, J.R.; Muhlrad, D.; Parker, R. P bodies promote stress granule assembly in saccharomyces cerevisiae. J. Cell Biol. 2008, 183, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Brengues, M.; Teixeira, D.; Parker, R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science 2005, 310, 486–489. [Google Scholar] [CrossRef] [PubMed]
- Teixeira, D.; Sheth, U.; Valencia-Sanchez, M.A.; Brengues, M.; Parker, R. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 2005, 11, 371–382. [Google Scholar] [CrossRef] [PubMed]
- Clemens, K.; Bilanchone, V.; Beliakova-Bethell, N.; Larsen, L.S.; Nguyen, K.; Sandmeyer, S. Sequence requirements for localization and packaging of Ty3 retroelement RNA. Virus Res. 2013, 171, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Bilanchone, V.; Clemens, K.; Kaake, R.; Dawson, A.R.; Matheos, D.; Nagashima, K.; Sitlani, P.; Patterson, K.; Chang, I.; Huang, L.; et al. Ty3 retrotransposon hijacks mating yeast RNA processing bodies to infect new genomes. PLoS Genet. 2015, 11, e1005528. [Google Scholar] [CrossRef] [PubMed]
- Dutko, J.A.; Kenny, A.E.; Gamache, E.R.; Curcio, M.J. 5′ to 3′ mRNA decay factors colocalize with Ty1 gag and human APOBEC3G and promote Ty1 retrotransposition. J. Virol. 2010, 84, 5052–5066. [Google Scholar] [CrossRef] [PubMed]
- Irwin, B.; Aye, M.; Baldi, P.; Beliakova-Bethell, N.; Cheng, H.; Dou, Y.; Liou, W.; Sandmeyer, S. Retroviruses and yeast retrotransposons use overlapping sets of host genes. Genome Res. 2005, 15, 641–654. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.L.; Coleman, L.E.; Raymond, A.S.; Goodson, S.G.; Pittard, W.S.; Tsui, C.; Devine, S.E. Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 2003, 164, 867–879. [Google Scholar] [PubMed]
- Mellor, J.; Malim, M.H.; Gull, K.; Tuite, M.F.; McCready, S.; Dibbayawan, T.; Kingsman, S.M.; Kingsman, A.J. Reverse transcriptase activity and Ty RNA are associated with virus-like particles in yeast. Nature 1985, 318, 583–586. [Google Scholar] [CrossRef] [PubMed]
- Luschnig, C.; Bachmair, A. RNA packaging of yeast retrotransposon Ty1 in the heterologous host, Escherichia coli. Biol. Chem. 1997, 378, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Summers, M.F. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011, 410, 609–633. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, V.; Summers, M.F. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Kutluay, S.B.; Zang, T.; Blanco-Melo, D.; Powell, C.; Jannain, D.; Errando, M.; Bieniasz, P.D. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Rein, A.; Datta, S.A.; Jones, C.P.; Musier-Forsyth, K. Diverse interactions of retroviral gag proteins with RNAs. Trends Biochem. Sci. 2011, 36, 373–380. [Google Scholar] [CrossRef] [PubMed]
- Purzycka, K.J.; Garfinkel, D.J.; Boeke, J.D.; le Grice, S.F. Influence of RNA structural elements on Ty1 retrotransposition. Mob. Genet. Elem. 2013. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.H.; Levin, H.L. Reverse transcription of a self-primed retrotransposon requires an RNA structure similar to the U5-IR stem-loop of retroviruses. Mol. Cell. Biol. 1998, 18, 6859–6869. [Google Scholar] [CrossRef] [PubMed]
- Shehu-Xhilaga, M.; Kraeusslich, H.G.; Pettit, S.; Swanstrom, R.; Lee, J.Y.; Marshall, J.A.; Crowe, S.M.; Mak, J. Proteolytic processing of the p2/nucleocapsid cleavage site is critical for human immunodeficiency virus type 1 RNA dimer maturation. J. Virol. 2001, 75, 9156–9164. [Google Scholar] [CrossRef] [PubMed]
- Kirchner, J.; Sandmeyer, S. Proteolytic processing of Ty3 proteins is required for transposition. J. Virol. 1993, 67, 19–28. [Google Scholar] [PubMed]
- Kuznetsov, Y.G.; Zhang, M.; Menees, T.M.; McPherson, A.; Sandmeyer, S. Investigation by atomic force microscopy of the structure of Ty3 retrotransposon particles. J. Virol. 2005, 79, 8032–8045. [Google Scholar] [CrossRef] [PubMed]
- Muller, F.; Bruhl, K.H.; Freidel, K.; Kowallik, K.V.; Ciriacy, M. Processing of Ty1 proteins and formation of Ty1 virus-like particles in saccharomyces cerevisiae. Mol. Gen. Genet. 1987, 207, 421–429. [Google Scholar] [CrossRef] [PubMed]
- Burns, N.R.; Saibil, H.R.; White, N.S.; Pardon, J.F.; Timmins, P.A.; Richardson, S.M.; Richards, B.M.; Adams, S.E.; Kingsman, S.M.; Kingsman, A.J. Symmetry, flexibility and permeability in the structure of yeast retrotransposon virus-like particles. EMBO J. 1992, 11, 1155–1164. [Google Scholar] [PubMed]
- Palmer, K.J.; Tichelaar, W.; Myers, N.; Burns, N.R.; Butcher, S.J.; Kingsman, A.J.; Fuller, S.D.; Saibil, H.R. Cryo-electron microscopy structure of yeast Ty retrotransposon virus-like particles. J. Virol. 1997, 71, 6863–6868. [Google Scholar] [PubMed]
- Luschnig, C.; Hess, M.; Pusch, O.; Brookman, J.; Bachmair, A. The gag homologue of retrotransposon Ty1 assembles into spherical particles in Escherichia coli. Eur. J. Biochem. 1995, 228, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Brookman, J.L.; Stott, A.J.; Cheeseman, P.J.; Adamson, C.S.; Holmes, D.; Cole, J.; Burns, N.R. Analysis of TYA protein regions necessary for formation of the Ty1 virus-like particle structure. Virology 1995, 212, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Martin-Rendon, E.; Marfany, G.; Wilson, S.; Ferguson, D.J.P.; Kingsman, S.M.; Kingsman, A.J. Structural determinants within the subunit protein of Ty1 virus-like particles. Mol. Microbiol. 1996, 22, 667–679. [Google Scholar] [CrossRef] [PubMed]
- Brookman, J.L.; Stott, A.J.; Cheeseman, P.J.; Burns, N.R.; Adams, S.E.; Kingsman, A.J.; Gull, K. An immunological analysis of Ty1 virus-like particle structure. Virology 1995, 207, 59–67. [Google Scholar] [CrossRef] [PubMed]
- Ganser-Pornillos, B.K.; Yeager, M.; Pornillos, O. Assembly and architecture of HIV. Adv. Exp. Med. Biol. 2012, 726, 441–465. [Google Scholar] [PubMed]
- Orlinsky, K.J.; Gu, J.; Hoyt, M.; Sandmeyer, S.; Menees, T.M. Mutations in the Ty3 major homology region affect multiple steps in Ty3 retrotransposition. J. Virol. 1996, 70, 3440–3448. [Google Scholar] [PubMed]
- Zhang, M.; Larsen, L.S.; Irwin, B.; Bilanchone, V.; Sandmeyer, S. Two-hybrid analysis of Ty3 capsid subdomain interactions. Mob. DNA 2010. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.W.; Noonan, K.; Aldovini, A. Nucleocapsid-RNA interactions are essential to structural stability but not to assembly of retroviruses. J. Virol. 2004, 78, 716–723. [Google Scholar] [CrossRef] [PubMed]
- Garfinkel, D.J.; Nyswaner, K.; Wang, J.; Cho, J.Y. Post-transcriptional cosuppression of Ty1 retrotransposition. Genetics 2003, 165, 83–99. [Google Scholar] [PubMed]
- Garfinkel, D.J.; Tucker, J.M.; Saha, A.; Nishida, Y.; Pachulska-Wieczorek, K.; Błaszczyk, L.; Purzycka, K.J. A self-encoded capsid derivative restricts Ty1 retrotransposition in Saccharomyces. Curr. Genet. 2016, 62, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Tucker, J.M.; Larango, M.E.; Wachsmuth, L.P.; Kannan, N.; Garfinkel, D.J. The Ty1 retrotransposon restriction factor p22 targets Gag. PLoS Genet. 2015, 11, e1005571. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pachulska-Wieczorek, K.; Le Grice, S.F.J.; Purzycka, K.J. Determinants of Genomic RNA Encapsidation in the Saccharomyces cerevisiae Long Terminal Repeat Retrotransposons Ty1 and Ty3. Viruses 2016, 8, 193. https://doi.org/10.3390/v8070193
Pachulska-Wieczorek K, Le Grice SFJ, Purzycka KJ. Determinants of Genomic RNA Encapsidation in the Saccharomyces cerevisiae Long Terminal Repeat Retrotransposons Ty1 and Ty3. Viruses. 2016; 8(7):193. https://doi.org/10.3390/v8070193
Chicago/Turabian StylePachulska-Wieczorek, Katarzyna, Stuart F.J. Le Grice, and Katarzyna J. Purzycka. 2016. "Determinants of Genomic RNA Encapsidation in the Saccharomyces cerevisiae Long Terminal Repeat Retrotransposons Ty1 and Ty3" Viruses 8, no. 7: 193. https://doi.org/10.3390/v8070193
APA StylePachulska-Wieczorek, K., Le Grice, S. F. J., & Purzycka, K. J. (2016). Determinants of Genomic RNA Encapsidation in the Saccharomyces cerevisiae Long Terminal Repeat Retrotransposons Ty1 and Ty3. Viruses, 8(7), 193. https://doi.org/10.3390/v8070193