The Life-Cycle of the HIV-1 Gag–RNA Complex
Abstract
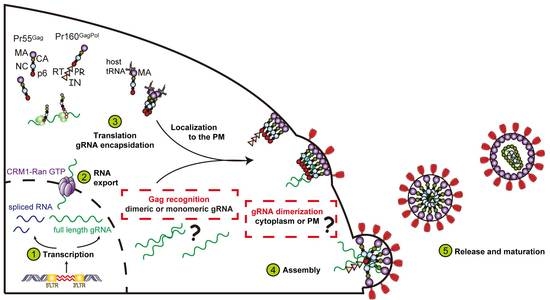
:1. Introduction
2. Nuclear Export of Genomic RNA and Its Role in Particle Assembly
3. The Fate of Genomic RNA: Translation versus Packaging
4. Selection of the Full-Length Genome by Gag
4.1. Gag
4.2. Genomic RNA and the Packaging Signal
4.3. Discrimination between Spliced and Genomic RNA, and the Regulation of Gag Binding
4.4. The Role of Cellular RNAs
5. Where Does the Gag–RNA Interaction Occur?
6. Transport of the Gag–RNA Complexes to Assembly Sites
7. Maturation
8. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ono, A. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol. Cell 2010, 102, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.G.; Summers, M.F. Structural biology of HIV. J. Mol. Biol. 1999, 285, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Sundquist, W.I.; Kräusslich, H.-G. HIV-1 assembly, budding, and maturation. Cold Spring Harb. Perspect. Med. 2012, 2, a006924. [Google Scholar] [CrossRef] [PubMed]
- Carlson, L.-A.; Bai, Y.; Keane, S.C.; Doudna, J.A.; Hurley, J.H. Reconstitution of selective HIV-1 RNA packaging in vitro by membrane-bound Gag assemblies. Elife 2016, 5, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Freed, E.O. HIV-1 assembly, release and maturation. Nat. Rev. Microbiol. 2015, 13, 484–496. [Google Scholar] [CrossRef] [PubMed]
- Burniston, M.T.; Cimarelli, A.; Colgan, J.; Curtis, S.P.; Luban, J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J. Virol. 1999, 73, 8527–8540. [Google Scholar] [PubMed]
- Muriaux, D.; Mirro, J.; Harvin, D.; Rein, A. RNA is a structural element in retrovirus particles. Proc. Natl. Acad. Sci. USA 2001, 98, 5246–5251. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Hu, W.S. HIV-1 RNA dimerization: It takes two to tango. AIDS Rev. 2009, 11, 91–102. [Google Scholar] [PubMed]
- Paillart, J.-C.; Shehu-Xhilaga, M.; Marquet, R.; Mak, J. Dimerization of retroviral RNA genomes: An inseparable pair. Nat. Rev. Microbiol. 2004, 2, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Kuzembayeva, M.; Dilley, K.; Sardo, L.; Hu, W.S. Life of psi: How full-length HIV-1 RNAs become packaged genomes in the viral particles. Virology 2014, 454–455, 362–370. [Google Scholar] [CrossRef] [PubMed]
- Olson, E.D.; Cantara, W.A.; Musier-Forsyth, K. New structure sheds light on selective HIV-1 genomic RNA packaging. Viruses 2015, 7, 4826–4835. [Google Scholar] [CrossRef] [PubMed]
- Abbink, T.E.M.; Berkhout, B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003, 278, 11601–11611. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Wahab, E.W.; Smyth, R.P.; Mailler, E.; Bernacchi, S.; Vivet-Boudou, V.; Hijnen, M.; Jossinet, F.; Mak, J.; Paillart, J.-C.; Marquet, R. Specific recognition of the HIV-1 genomic RNA by the Gag precursor. Nat. Commun. 2014, 5, 4304. [Google Scholar] [CrossRef] [PubMed]
- Van Bel, N.; Das, A.T.; Cornelissen, M.T.E.; Abbink, T.E.M.; Berkhout, B. A short sequence motif in the 5′ leader of the HIV-1 genome modulates extended RNA dimer formation and virus replication. J. Biol. Chem. 2014, 289, 35061–35074. [Google Scholar] [CrossRef] [PubMed]
- Keane, S.C.; Heng, X.; Lu, K.; Kharytonchyk, S.; Ramakrishnan, V.; Carter, G.; Barton, S.; Hosic, A.; Florwick, A.; Santos, J.; et al. RNA structure. Structure of the HIV-1 RNA packaging signal. Science 2015, 348, 917–921. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Heng, X.; Garyu, L.; Monti, S.; Garcia, E.L.; Kharytonchyk, S.; Dorjsuren, B.; Kulandaivel, G.; Jones, S.; Hiremath, A.; et al. NMR detection of structures in the HIV-1 5′-leader RNA that regulate genome packaging. Science 2011, 334, 242–245. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, R.D.; Luban, J.; Goff, S.P. Specific binding of human immunodeficiency virus type 1 Gag polyprotein and nucleocapsid protein to viral RNAs detected by RNA mobility shift assays. J. Virol. 1993, 67, 7190–7200. [Google Scholar] [PubMed]
- Lu, K.; Heng, X.; Summers, M.F. Structural determinants and mechanism of HIV-1 genome packaging. J. Mol. Biol. 2011, 410, 609–633. [Google Scholar] [CrossRef] [PubMed]
- Luban, J.; Goff, S.P. Binding of human immunodeficiency virus type 1 (HIV-1) RNA to recombinant HIV-1 Gag polyprotein. J. Virol. 1991, 65, 3203–3212. [Google Scholar] [PubMed]
- Luban, J.; Goff, S.P. Mutational analysis of cis-acting packaging signals in human immunodeficiency virus type 1 RNA. J. Virol. 1994, 68, 3784–3793. [Google Scholar] [PubMed]
- McBride, M.S.; Panganiban, A.T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 1996, 70, 2963–2973. [Google Scholar] [PubMed]
- Lever, A.; Gottlinger, H.; Haseltine, W.; Sodroski, J. Identification of a sequence required for efficient packaging of human immunodeficiency virus type 1 RNA into virions. J. Virol. 1989, 63, 4085–4087. [Google Scholar] [PubMed]
- Aldovini, A.; Young, R.A. Mutations of RNA and protein sequences involved in human immunodeficiency virus type 1 packaging result in production of noninfectious virus. J. Virol. 1990, 64, 1920–1926. [Google Scholar] [PubMed]
- Chamanian, M.; Purzycka, K.J.; Wille, P.T.; Ha, J.S.; McDonald, D.; Gao, Y.; Le Grice, S.F.J.; Arts, E.J. A cis-acting element in retroviral genomic RNA links Gag-Pol ribosomal frameshifting to selective viral RNA encapsidation. Cell Host Microbe 2013, 13, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Grewe, B.; Ehrhardt, K.; Hoffmann, B.; Blissenbach, M.; Brandt, S.; Uberla, K. The HIV-1 Rev protein enhances encapsidation of unspliced and spliced, RRE-containing lentiviral vector RNA. PLoS ONE 2012, 7, e48688. [Google Scholar] [CrossRef] [PubMed]
- Brandt, S.; Blissenbach, M.; Grewe, B.; Konietzny, R.; Grunwald, T.; Uberla, K. Rev proteins of human and simian immunodeficiency virus enhance RNA encapsidation. PLoS Pathog. 2007, 3, e54. [Google Scholar] [CrossRef] [PubMed]
- Purcell, D.F.; Martin, M.A. Alternative splicing of human immunodeficiency virus type 1 mRNA modulates viral protein expression, replication, and infectivity. J. Virol. 1993, 67, 6365–6378. [Google Scholar] [PubMed]
- Malim, M.H.; Hauber, J.; Le, S.Y.; Maizel, J.V.; Cullen, B.R. The HIV-1 Rev trans-activator acts through a structured target sequence to activate nuclear export of unspliced viral mRNA. Nature 1989, 338, 254–257. [Google Scholar] [CrossRef] [PubMed]
- Felber, B.K.; Hadzopoulou-Cladaras, M.; Cladaras, C.; Copeland, T.; Pavlakis, G.N. Rev protein of human immunodeficiency virus type 1 affects the stability and transport of the viral mRNA. Proc. Natl. Acad. Sci. USA 1989, 86, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Daly, T.J.; Cook, K.S.; Gray, G.S.; Maione, T.E.; Rusche, J.R. Specific binding of HIV-1 recombinant Rev protein to the Rev-responsive element in vitro. Nature 1989, 342, 816–819. [Google Scholar] [CrossRef] [PubMed]
- Wen, W.; Meinkoth, J.L.; Tsien, R.Y.; Taylor, S.S. Identification of a signal for rapid export of proteins from the nucleus. Cell 1995, 82, 463–473. [Google Scholar] [CrossRef]
- Fornerod, M.; Ohno, M.; Yoshida, M.; Mattaj, I.W. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 1997, 90, 1051–1060. [Google Scholar] [CrossRef]
- Pollard, V.W.; Malim, M.H. The HIV-1 Rev protein. Annu. Rev. Microbiol. 1998, 52, 491–532. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Sturgeon, T.; Chen, C.; Watkins, S.C.; Weisz, O.A.; Montelaro, R.C. Distinct intracellular trafficking of equine infectious anemia virus and human immunodeficiency virus type 1 Gag during viral assembly and budding revealed by bimolecular fluorescence complementation assays. J. Virol. 2007, 81, 11226–11235. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Sturgeon, T.; Weisz, O.A.; Mothes, W.; Montelaro, R.C. HIV-1 matrix dependent membrane targeting is regulated by Gag mRNA trafficking. PLoS ONE 2009, 4, e6551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mariani, R.; Rutter, G.; Harris, M.E.; Hope, T.J.; Kräusslich, H.G.; Landau, N.R. A block to human immunodeficiency virus type 1 assembly in murine cells. J. Virol. 2000, 74, 3859–3870. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.K.; Rousso, I.; Shim, S.; Kim, P.S. Efficient assembly of an HIV-1/MLV Gag-chimeric virus in murine cells. Proc. Natl. Acad. Sci. USA 2001, 98, 15239–15244. [Google Scholar] [CrossRef] [PubMed]
- Sherer, N.M.; Swanson, C.M.; Papaioannou, S.; Malim, M.H. Matrix mediates the functional link between human immunodeficiency virus type 1 RNA nuclear export elements and the assembly competency of Gag in murine cells. J. Virol. 2009, 83, 8525–8535. [Google Scholar] [CrossRef] [PubMed]
- Swanson, C.M.; Puffer, B.A.; Ahmad, K.M.; Doms, R.W.; Malim, M.H. Retroviral mRNA nuclear export elements regulate protein function and virion assembly. EMBO J. 2004, 23, 2632–2640. [Google Scholar] [CrossRef] [PubMed]
- Hatziioannou, T.; Martin-Serrano, J.; Zang, T.; Bieniasz, P.D. Matrix-induced inhibition of membrane binding contributes to human immunodeficiency virus type 1 particle assembly defects in murine cells. J. Virol. 2005, 79, 15586–15589. [Google Scholar] [CrossRef] [PubMed]
- Pocock, G.M.; Becker, J.T.; Swanson, C.M.; Ahlquist, P.; Sherer, N.M. HIV-1 and M-PMV RNA nuclear export elements program viral genomes for distinct cytoplasmic trafficking Behaviors. PLoS Pathog. 2016, 12, e1005565. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Grunwald, D.; Sardo, L.; Galli, A.; Plisov, S.; Nikolaitchik, O.A.; Chen, D.; Lockett, S.; Larson, D.R.; Pathak, V.K.; et al. Cytoplasmic HIV-1 RNA is mainly transported by diffusion in the presence or absence of Gag protein. Proc. Natl. Acad. Sci. USA 2014, 111, E5205–E5213. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.G.; Rosenak, M.J. Synthesis of murine leukemia virus proteins associated with virions assembled in actinomycin D-treated cells: Evidence for persistence of viral messenger RNA. Proc. Natl. Acad. Sci. USA 1976, 73, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Messer, L.I.; Levin, J.G.; Chattopadhyay, S.K. Metabolism of viral RNA in murine leukemia virus-infected cells; evidence for differential stability of viral message and virion precursor RNA. J. Virol. 1981, 40, 683–690. [Google Scholar] [PubMed]
- Dorman, N.; Lever, A. Comparison of viral genomic RNA sorting mechanisms in human immunodeficiency virus type 1 (HIV-1), HIV-2, and Moloney murine leukemia virus. J. Virol. 2000, 74, 11413–11417. [Google Scholar] [CrossRef] [PubMed]
- Griffin, S.D.; Allen, J.F.; Lever, A.M. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: Cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 2001, 75, 12058–12069. [Google Scholar] [CrossRef] [PubMed]
- Kaye, J.F.; Lever, A.M. Human immunodeficiency virus types 1 and 2 differ in the predominant mechanism used for selection of genomic RNA for encapsidation. J. Virol. 1999, 73, 3023–3031. [Google Scholar] [PubMed]
- Butsch, M.; Boris-Lawrie, K. Translation is not required To generate virion precursor RNA in human immunodeficiency virus type 1-infected T cells. J. Virol. 2000, 74, 11531–11537. [Google Scholar] [CrossRef] [PubMed]
- Poon, D.T.K.; Chertova, E.N.; Ott, D.E. Human immunodeficiency virus type 1 preferentially encapsidates genomic RNAs that encode Pr55(Gag): Functional linkage between translation and RNA packaging. Virology 2002, 293, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Hu, J.; Russell, R.S.; Wainberg, M.A. Translation of Pr55(Gag) augments packaging of human immunodeficiency virus type 1 RNA in a cis-acting manner. AIDS Res. Hum. Retroviruses 2002, 18, 1117–1126. [Google Scholar] [CrossRef] [PubMed]
- Nikolaitchik, O.A.; Rhodes, T.D.; Ott, D.; Hu, W.-S. Effects of mutations in the human immunodeficiency virus type 1 Gag gene on RNA packaging and recombination. J. Virol. 2006, 80, 4691–4697. [Google Scholar] [CrossRef] [PubMed]
- Ni, N.; Nikolaitchik, O.A.; Dilley, K.A.; Chen, J.; Galli, A.; Fu, W.; Prasad, V.V.S.P.; Ptak, R.G.; Pathak, V.K.; Hu, W.-S. Mechanisms of human immunodeficiency virus type 2 RNA packaging: Efficient trans packaging and selection of RNA copackaging partners. J. Virol. 2011, 85, 7603–7612. [Google Scholar] [CrossRef] [PubMed]
- Olety, B.; Ono, A. Roles played by acidic lipids in HIV-1 Gag membrane binding. Virus Res. 2014, 193, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Alfadhli, A.; Still, A.; Barklis, E. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J. Virol. 2009, 83, 12196–12203. [Google Scholar] [CrossRef] [PubMed]
- Chukkapalli, V.; Oh, S.J.; Ono, A. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc. Natl. Acad. Sci. USA 2010, 107, 1600–1605. [Google Scholar] [CrossRef] [PubMed]
- Alfadhli, A.; McNett, H.; Tsagli, S.; Bächinger, H.P.; Peyton, D.H.; Barklis, E. HIV-1 matrix protein binding to RNA. J. Mol. Biol. 2011, 410, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Kutluay, S.B.; Zang, T.; Blanco-melo, D.; Powell, C.; Jannain, D.; Errando, M. Global changes in the RNA binding specificity of HIV-1 Gag regulate virion genesis. Cell 2014, 159, 1096–1109. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Rein, A. In vitro assembly properties of human immunodeficiency virus type 1 Gag protein lacking the p6 domain. J. Virol. 1999, 73, 2270–2279. [Google Scholar] [PubMed]
- Cimarelli, A.; Sandin, S.; Höglund, S.; Luban, J. Basic residues in human immunodeficiency virus type 1 nucleocapsid promote virion assembly via interaction with RNA. J. Virol. 2000, 74, 3046–3057. [Google Scholar] [CrossRef] [PubMed]
- Cruceanu, M.; Urbaneja, M.A.; Hixson, C.V.; Johnson, D.G.; Datta, S.A.; Fivash, M.J.; Stephen, A.G.; Fisher, R.J.; Gorelick, R.J.; Casas-Finet, J.R.; et al. Nucleic acid binding and chaperone properties of HIV-1 Gag and nucleocapsid proteins. Nucleic Acids Res. 2006, 34, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Damgaard, C.K.; Dyhr-Mikkelsen, H.; Kjems, J. Mapping the RNA binding sites for human immunodeficiency virus type-1 Gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 1998, 26, 3667–3676. [Google Scholar] [CrossRef] [PubMed]
- Rein, A.; Datta, S.A.K.; Jones, C.P.; Musier-Forsyth, K. Diverse interactions of retroviral Gag proteins with RNAs. Trends Biochem. Sci. 2011, 36, 373–380. [Google Scholar] [PubMed]
- Crist, R.M.; Datta, S.A.K.; Stephen, A.G.; Soheilian, F.; Mirro, J.; Fisher, R.J.; Nagashima, K.; Rein, A. Assembly properties of human immunodeficiency virus type 1 Gag-leucine zipper chimeras: Implications for retrovirus assembly. J. Virol. 2009, 83, 2216–2225. [Google Scholar] [CrossRef] [PubMed]
- Klein, K.C.; Reed, J.C.; Tanaka, M.; Nguyen, V.T.; Giri, S.; Lingappa, J.R. HIV Gag-leucine zipper chimeras form ABCE1-containing intermediates and RNase-resistant immature capsids similar to those formed by wild-type HIV-1 Gag. J. Virol. 2011, 85, 7419–7435. [Google Scholar] [CrossRef] [PubMed]
- Mattei, S.; Flemming, A.; Anders-Össwein, M.; Kräusslich, H.-G.; Briggs, J.A.G.; Müller, B. RNA and nucleocapsid are dispensable for mature HIV-1 capsid assembly. J. Virol. 2015, 89, 9739–9747. [Google Scholar] [CrossRef] [PubMed]
- Accola, M.A.; Strack, B.; Göttlinger, H.G. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 2000, 74, 5395–5402. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qian, H.; Love, Z.; Barklis, E. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J. Virol. 1998, 72, 1782–1789. [Google Scholar] [PubMed]
- Smyth, R.P.; Despons, L.; Huili, G.; Bernacchi, S.; Hijnen, M.; Mak, J.; Jossinet, F.; Weixi, L.; Paillart, J.; von Kleist, M.; et al. Mutational interference mapping experiment (MIME) for studying RNA structure and function. Nat. Methods 2015, 12, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Nikolaitchik, O.A.; Singh, J.; Wright, A.; Bencsics, C.E.; Coffin, J.M.; Ni, N.; Lockett, S.; Pathak, V.K.; Hu, W.-S. High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. USA 2009, 106, 13535–13540. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.C.; Marquet, R.; Skripkin, E.; Ehresmann, B.; Ehresmann, C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J. Biol. Chem. 1994, 269, 27486–27493. [Google Scholar] [PubMed]
- Paillart, J.C.; Skripkin, E.; Ehresmann, B.; Ehresmann, C.; Marquet, R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. USA 1996, 93, 5572–5577. [Google Scholar] [CrossRef] [PubMed]
- Skripkin, E.; Paillart, J.C.; Marquet, R.; Ehresmann, B.; Ehresmann, C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl. Acad. Sci. USA 1994, 91, 4945–4949. [Google Scholar] [CrossRef] [PubMed]
- Skripkin, E.; Paillart, J.C.; Marquet, R.; Blumenfeld, M.; Ehresmann, B.; Ehresmann, C. Mechanisms of inhibition of in vitro dimerization of HIV type I RNA by sense and antisense oligonucleotides. J. Biol. Chem. 1996, 271, 28812–28817. [Google Scholar] [PubMed]
- Jones, K.L.; Sonza, S.; Mak, J. Primary T-lymphocytes rescue the replication of HIV-1 DIS RNA mutants in part by facilitating reverse transcription. Nucleic Acids Res. 2008, 36, 1578–1588. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.C.; Berthoux, L.; Ottmann, M.; Darlix, J.L.; Marquet, R.; Ehresmann, B.; Ehresmann, C. A dual role of the putative RNA dimerization initiation site of human immunodeficiency virus type 1 in genomic RNA packaging and proviral DNA synthesis. J. Virol. 1996, 70, 8348–8354. [Google Scholar] [PubMed]
- Houzet, L.; Paillart, J.C.; Smagulova, F.; Maurel, S.; Morichaud, Z.; Marquet, R.; Mougel, M. HIV controls the selective packaging of genomic, spliced viral and cellular RNAs into virions through different mechanisms. Nucleic Acids Res. 2007, 35, 2695–2704. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, M.; Fay, P.J.; Bambara, R.A. The kissing hairpin sequence promotes recombination within the HIV-I 5′ leader region. J. Biol. Chem. 2001, 276, 36482–36492. [Google Scholar] [CrossRef] [PubMed]
- Smyth, R.P.; Davenport, M.P.; Mak, J. The origin of genetic diversity in HIV-1. Virus Res. 2012, 169, 415–429. [Google Scholar] [CrossRef] [PubMed]
- Smyth, R.P.; Schlub, T.E.; Grimm, A.J.; Waugh, C.; Ellenberg, P.; Chopra, A.; Mallal, S.; Cromer, D.; Mak, J.; Davenport, M.P. Identifying Recombination Hot Spots in the HIV-1 Genome. J. Virol. 2014, 88, 2891–2902. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Kafaie, J.; Yang, L.; Laughrea, M. HIV-1 viral RNA is selected in the form of monomers that dimerize in a three-step protease-dependent process; the DIS of stem-loop 1 initiates viral RNA dimerization. J. Mol. Biol. 2007, 371, 1084–1098. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.K.; Shehu-Xhilaga, M.; Campbell, S.M.; Poumbourios, P.; Crowe, S.M.; Mak, J. The dimer initiation sequence stem-loop of human immunodeficiency virus type 1 is dispensable for viral replication in peripheral blood mononuclear cells. J. Virol. 2003, 77, 8329–8335. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.D.; Fu, W.; Nikolaitchik, O.A.; Chen, J.; Ptak, R.G.; Hu, W.-S. Dimer initiation signal of human immunodeficiency virus type 1: Its role in partner selection during RNA copackaging and its effects on recombination. J. Virol. 2007, 81, 4002–4011. [Google Scholar] [CrossRef] [PubMed]
- Shen, N.; Jetté, L.; Liang, C.; Wainberg, M.A.; Laughrea, M. Impact of human immunodeficiency virus type 1 RNA dimerization on viral infectivity and of stem-loop B on RNA dimerization and reverse transcription and dissociation of dimerization from packaging. J. Virol. 2000, 74, 5729–5735. [Google Scholar] [CrossRef] [PubMed]
- Berkhout, B.; van Wamel, J.L. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 1996, 70, 6723–6732. [Google Scholar] [PubMed]
- Sakuragi, J.I.; Panganiban, A.T. Human immunodeficiency virus type 1 RNA outside the primary encapsidation and dimer linkage region affects RNA dimer stability in vivo. J. Virol. 1997, 71, 3250–3254. [Google Scholar] [PubMed]
- Harrison, G.P.; Lever, A.M. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J. Virol. 1992, 66, 4144–4153. [Google Scholar] [PubMed]
- Harrison, G.P.; Miele, G.; Hunter, E.; Lever, A.M. Functional analysis of the core human immunodeficiency virus type 1 packaging signal in a permissive cell line. J. Virol. 1998, 72, 5886–5896. [Google Scholar] [PubMed]
- Paillart, J.C.; Dettenhofer, M.; Yu, X.F.; Ehresmann, C.; Ehresmann, B.; Marquet, R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004, 279, 48397–48403. [Google Scholar] [CrossRef] [PubMed]
- Siegfried, N.A.; Busan, S.; Rice, G.M.; Nelson, J.A.E.; Weeks, K.M. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat. Methods 2014, 11, 959–965. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.M.; Dang, K.K.; Gorelick, R.J.; Leonard, C.W.; Bess, J.W., Jr.; Swanstrom, R.; Burch, C.L.; Weeks, K.M. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature 2009, 460, 711–716. [Google Scholar] [CrossRef] [PubMed]
- Clavel, F.; Orenstein, J.M. A mutant of human immunodeficiency virus with reduced RNA packaging and abnormal particle morphology. J. Virol. 1990, 64, 5230–5234. [Google Scholar] [PubMed]
- D’Souza, V.; Summers, M.F. How retroviruses select their genomes. Nat. Rev. Microbiol. 2005, 3, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Berkowitz, R.; Fisher, J.; Goff, S.P. RNA packaging. Curr. Top. Microbiol. Immunol. 1996, 214, 177–218. [Google Scholar] [PubMed]
- Helga-Maria, C.; Hammarskjöld, M.L.; Rekosh, D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J. Virol. 1999, 73, 4127–4135. [Google Scholar] [PubMed]
- Russell, R.S.; Hu, J.; Laughrea, M.; Wainberg, M.A.; Liang, C. Deficient dimerization of human immunodeficiency virus type 1 RNA caused by mutations of the u5 RNA sequences. Virology 2002, 303, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Clever, J.L.; Miranda, D.; Parslow, T.G.; Miranda, D., Jr.; Parslow, T.G. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J. Virol. 2002, 76, 12381–12387. [Google Scholar] [CrossRef] [PubMed]
- Laughrea, M.; Jetté, L.; Mak, J.; Kleiman, L.; Liang, C.; Wainberg, M.A. Mutations in the kissing-loop hairpin of human immunodeficiency virus type 1 reduce viral infectivity as well as genomic RNA packaging and dimerization. J. Virol. 1997, 71, 3397–3406. [Google Scholar] [PubMed]
- Göttlinger, H.G.; Dorfman, T.; Sodroski, J.G.; Haseltine, W.A. Effect of mutations affecting the p6 Gag protein on human immunodeficiency virus particle release. Proc. Natl. Acad. Sci. USA 1991, 88, 3195–3199. [Google Scholar] [CrossRef] [PubMed]
- Huang, M.; Orenstein, J.M.; Martin, M.A.; Freed, E.O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J. Virol. 1995, 69, 6810–6818. [Google Scholar] [PubMed]
- Demirov, D.G.; Orenstein, J.M.; Freed, E.O. The late domain of human immunodeficiency virus type 1 p6 promotes virus release in a cell type-dependent manner. J. Virol. 2002, 76, 105–117. [Google Scholar] [CrossRef] [PubMed]
- McKinstry, W.J.; Hijnen, M.; Tanwar, H.S.; Sparrow, L.G.; Nagarajan, S.; Pham, S.T.; Mak, J. Expression and purification of soluble recombinant full length HIV-1 Pr55(Gag) protein in Escherichia coli. Protein Expr. Purif. 2014, 100, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, K.A.; Gorelick, R.J.; Vasa, S.M.; Guex, N.; Rein, A.; Mathews, D.H.; Giddings, M.C.; Weeks, K.M. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008, 6, e96. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, J.C.; Prestwood, L.J.; Lever, A.M.L. A novel combined RNA-protein interaction analysis distinguishes HIV-1 Gag protein binding sites from structural change in the viral RNA leader. Sci. Rep. 2015, 5, 14369. [Google Scholar] [CrossRef] [PubMed]
- Harrich, D.; Hooker, C.W.; Parry, E. The human immunodeficiency virus type 1 TAR RNA upper stem-loop plays distinct roles in reverse transcription and RNA packaging. J. Virol. 2000, 74, 5639–5646. [Google Scholar] [CrossRef] [PubMed]
- McBride, M.S.; Schwartz, M.D.; Panganiban, A.T. Efficient encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J. Virol. 1997, 71, 4544–4554. [Google Scholar] [PubMed]
- Didierlaurent, L.; Racine, P.J.; Houzet, L.; Chamontin, C.; Berkhout, B.; Mougel, M. Role of HIV-1 RNA and protein determinants for the selective packaging of spliced and unspliced viral RNA and host U6 and 7SL RNA in virus particles. Nucleic Acids Res. 2011, 39, 8915–8927. [Google Scholar] [CrossRef] [PubMed]
- Nikolaitchik, O.A.; Hu, W.-S. Deciphering the role of the Gag-Pol ribosomal frameshift signal in HIV-1 RNA genome packaging. J. Virol. 2014, 88, 4040–4046. [Google Scholar] [CrossRef] [PubMed]
- Nikolaitchik, O.A.; Moore, M.D.; Chen, J.; Hammarskjöld, M.-L.; Rekosh, D.; Hu, W.-S. Probing the HIV-1 genomic RNA trafficking pathway and dimerization by genetic recombination and single virion analyses. PLoS Pathog. 2009, 5, e1000627. [Google Scholar]
- Sakuragi, J.; Shioda, T.; Panganiban, A.T. Duplication of the primary encapsidation and dimer linkage region of human immunodeficiency virus type 1 RNA results in the appearance of monomeric RNA in virions. J. Virol. 2001, 75, 2557–2565. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Rahman, S.A.; Nikolaitchik, O.A.; Grunwald, D.; Sardo, L.; Burdick, R.C.; Plisov, S.; Liang, E.; Tai, S.; Pathak, V.K.; et al. HIV-1 RNA genome dimerizes on the plasma membrane in the presence of Gag protein. Proc. Natl. Acad. Sci. USA 2016, 113, E201–E208. [Google Scholar] [CrossRef] [PubMed]
- Clever, J.L.; Parslow, T.G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J. Virol. 1997, 71, 3407–3414. [Google Scholar] [PubMed]
- Ooms, M.; Verhoef, K.; Southern, E.; Huthoff, H.; Berkhout, B. Probing alternative foldings of the HIV-1 leader RNA by antisense oligonucleotide scanning arrays. Nucleic Acids Res. 2004, 32, 819–827. [Google Scholar] [CrossRef] [PubMed]
- Paillart, J.C.; Marquet, R.; Skripkin, E.; Ehresmann, C.; Ehresmann, B. Dimerization of retroviral genomic RNAs: Structural and functional implications. Biochimie 1996, 78, 639–653. [Google Scholar] [CrossRef]
- Russell, R.S.; Liang, C.; Wainberg, M.A. Is HIV-1 RNA dimerization a prerequisite for packaging? Yes, no, probably? Retrovirology 2004, 1, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aktar, S.J.; Jabeen, A.; Ali, L.M.; Vivet-Boudou, V.; Marquet, R.; Rizvi, T.A. SHAPE analysis of the 5′ end of the Mason-Pfizer monkey virus (MPMV) genomic RNA reveals structural elements required for genome dimerization. RNA 2013, 19, 1648–1658. [Google Scholar] [CrossRef] [PubMed]
- Aktar, S.J.; Vivet-Boudou, V.; Ali, L.M.; Jabeen, A.; Kalloush, R.M.; Richer, D.; Mustafa, F.; Marquet, R.; Rizvi, T.A. Structural basis of genomic RNA (gRNA) dimerization and packaging determinants of mouse mammary tumor virus (MMTV). Retrovirology 2014, 11, 96. [Google Scholar] [CrossRef] [PubMed]
- Kalloush, R.M.; Vivet-Boudou, V.; Ali, L.M.; Mustafa, F.; Marquet, R.; Rizvi, T.A. Packaging of Mason-Pfizer monkey virus (MPMV) genomic RNA depends upon conserved long-range interactions (LRIs) between U5 and Gag sequences. RNA 2016, 22, 905–919. [Google Scholar] [CrossRef] [PubMed]
- Huthoff, H.; Berkhout, B. Two alternating structures of the HIV-1 leader RNA. RNA 2001, 7, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Miele, G.; Mouland, A.; Harrison, G.P.; Cohen, E.; Lever, A.M. The human immunodeficiency virus type 1 5′ packaging signal structure affects translation but does not function as an internal ribosome entry site structure. J. Virol. 1996, 70, 944–951. [Google Scholar] [PubMed]
- Damgaard, C.K.; Andersen, E.S.; Knudsen, B.; Gorodkin, J.; Kjems, J. RNA interactions in the 5′ region of the HIV-1 genome. J. Mol. Biol. 2004, 336, 369–379. [Google Scholar] [CrossRef] [PubMed]
- Tran, T.; Liu, Y.; Marchant, J.; Monti, S.; Seu, M.; Zaki, J.; Yang, A.L.; Bohn, J.; Ramakrishnan, V.; Singh, R.; et al. Conserved determinants of lentiviral genome dimerization. Retrovirology 2015, 12, 83. [Google Scholar] [CrossRef] [PubMed]
- Heng, X.; Kharytonchyk, S.; Garcia, E.L.; Lu, K.; Divakaruni, S.S.; Lacotti, C.; Edme, K.; Telesnitsky, A.; Summers, M.F. Identification of a Minimal Region of the HIV-1 5′-Leader Required for RNA Dimerization, NC Binding, and Packaging. J. Mol. Biol. 2012, 417, 224–239. [Google Scholar] [CrossRef] [PubMed]
- Song, R.; Kafaie, J.; Laughrea, M. Role of the 5′ TAR stem--loop and the U5-AUG duplex in dimerization of HIV-1 genomic RNA. Biochemistry 2008, 47, 3283–3293. [Google Scholar] [CrossRef] [PubMed]
- Eckwahl, M.J.; Arnion, H.; Kharytonchyk, S.; Zang, T.; Bieniasz, P.D.; Telesnitsky, A.; Wolin, S.L. Analysis of the human immunodeficiency virus-1 RNA packageome. RNA 2016, 22, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Muriaux, D.; Rein, A. Encapsidation and transduction of cellular genes by retroviruses. Front. Biosci. 2003, 8, d135–d142. [Google Scholar] [PubMed]
- Onafuwa-Nuga, A.A.; Telesnitsky, A.; King, S.R. 7SL RNA, but not the 54-kd signal recognition particle protein, is an abundant component of both infectious HIV-1 and minimal virus-like particles. RNA 2006, 12, 542–546. [Google Scholar] [CrossRef] [PubMed]
- Rulli, S.J.; Hibbert, C.S.; Mirro, J.; Pederson, T.; Biswal, S.; Rein, A. Selective and nonselective packaging of cellular RNAs in retrovirus particles. J. Virol. 2007, 81, 6623–6631. [Google Scholar] [CrossRef] [PubMed]
- Eckwahl, M.J.; Telesnitsky, A.; Wolin, S.L. Host RNA packaging by retroviruses: A newly synthesized story. mBio 2016, 7, e02025-15. [Google Scholar] [CrossRef] [PubMed]
- Telesnitsky, A.; Wolin, S.L. The host RNAs in retroviral particles. Viruses 2016, 8, 235. [Google Scholar] [CrossRef] [PubMed]
- Campbell, S.; Vogt, V.M. Self-assembly in vitro of purified CA-NC proteins from Rous sarcoma virus and human immunodeficiency virus type 1. J. Virol. 1995, 69, 6487–6497. [Google Scholar] [PubMed]
- Hendrix, J.; Baumgärtel, V.; Schrimpf, W.; Ivanchenko, S.; Digman, M.A.; Gratton, E.; Kräusslich, H.-G.; Müller, B.; Lamb, D.C. Live-cell observation of cytosolic HIV-1 assembly onset reveals RNA-interacting Gag oligomers. J. Cell Biol. 2015, 210, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Lainé, S.; Pessel-Vivares, L.; Mougel, M. Cell biology of retroviral RNA packaging. RNA Biol. 2011, 8, 572–580. [Google Scholar] [CrossRef] [PubMed]
- Garbitt-Hirst, R.; Kenney, S.P.; Parent, L.J. Genetic evidence for a connection between Rous sarcoma virus Gag nuclear trafficking and genomic RNA packaging. J. Virol. 2009, 83, 6790–6797. [Google Scholar] [CrossRef] [PubMed]
- Scheifele, L.Z.; Garbitt, R.A.; Rhoads, J.D.; Parent, L.J. Nuclear entry and CRM1-dependent nuclear export of the Rous sarcoma virus Gag polyprotein. Proc. Natl. Acad. Sci. USA 2002, 99, 3944–3949. [Google Scholar] [CrossRef] [PubMed]
- Gudleski, N.; Flanagan, J.M.; Ryan, E.P.; Bewley, M.C.; Parent, L.J. Directionality of nucleocytoplasmic transport of the retroviral Gag protein depends on sequential binding of karyopherins and viral RNA. Proc. Natl. Acad. Sci. USA 2010, 107, 9358–9363. [Google Scholar] [CrossRef] [PubMed]
- Stake, M.S.; Bann, D.V.; Kaddis, R.J.; Parent, L.J. Nuclear trafficking of retroviral RNAs and Gag proteins during late steps of replication. Viruses 2013, 5, 2767–2795. [Google Scholar] [CrossRef] [PubMed]
- Baluyot, M.F.; Grosse, S.A.; Lyddon, T.D.; Janaka, S.K.; Johnson, M.C. CRM1-dependent trafficking of retroviral Gag proteins revisited. J. Virol. 2012, 86, 4696–4700. [Google Scholar] [CrossRef] [PubMed]
- Lochmann, T.L.; Bann, D.V.; Ryan, E.P.; Beyer, A.R.; Mao, A.; Cochrane, A.; Parent, L.J. NC-mediated nucleolar localization of retroviral Gag proteins. Virus Res. 2013, 171, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Yu, K.L.; Lee, S.H.; Lee, E.S.; You, J.C. HIV-1 nucleocapsid protein localizes efficiently to the nucleus and nucleolus. Virology 2016, 492, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Grewe, B.; Hoffmann, B.; Ohs, I.; Blissenbach, M.; Brandt, S.; Tippler, B.; Grunwald, T.; Uberla, K. Cytoplasmic utilization of human immunodeficiency virus type 1 genomic RNA is not dependent on a nuclear interaction with Gag. J. Virol. 2012, 86, 2990–3002. [Google Scholar] [CrossRef] [PubMed]
- Ferrer, M.; Clerté, C.; Chamontin, C.; Basyuk, E.; Lainé, S.; Hottin, J.; Bertrand, E.; Margeat, E.; Mougel, M. Imaging HIV-1 RNA dimerization in cells by multicolor super-resolution and fluctuation microscopies. Nucleic Acids Res. 2016. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Bieniasz, P.D.; Simon, S.M. Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 2008, 454, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Kemler, I.; Meehan, A.; Poeschla, E.M. Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J. Virol. 2010, 84, 6352–6366. [Google Scholar] [CrossRef] [PubMed]
- Kutluay, S.B.; Bieniasz, P.D. Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog. 2010, 6, e1001200. [Google Scholar] [CrossRef] [PubMed]
- Poole, E.; Strappe, P.; Mok, H.-P.; Hicks, R.; Lever, A.M.L. HIV-1 Gag-RNA interaction occurs at a perinuclear/centrosomal site; analysis by confocal microscopy and FRET. Traffic 2005, 6, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Bernacchi, S. HIV-1 Pr55Gag binds genomic and spliced RNAs with different affinity and stoichiometry. 2016; unpublished. [Google Scholar]
- Ott, D.E.; Coren, L.V.; Kane, B.P.; Busch, L.K.; Johnson, D.G.; Sowder, R.C.; Chertova, E.N.; Arthur, L.O.; Henderson, L.E. Cytoskeletal proteins inside human immunodeficiency virus type 1 virions. J. Virol. 1996, 70, 7734–7743. [Google Scholar] [PubMed]
- Gladnikoff, M.; Shimoni, E.; Gov, N.S.; Rousso, I. Retroviral assembly and budding occur through an actin-driven mechanism. Biophys. J. 2009, 97, 2419–2428. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Neil, S.J.D.; Bess, C.; Johnson, M.C.; Virgen, C.A.; Simon, S.M.; Bieniasz, P.D. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol. 2006, 4, e435. [Google Scholar] [CrossRef] [PubMed]
- Rahman, S.A.; Koch, P.; Weichsel, J.; Godinez, W.J.; Schwarz, U.; Rohr, K.; Lamb, D.C.; Kräusslich, H.-G.; Müller, B. Investigating the role of F-actin in human immunodeficiency virus assembly by live-cell microscopy. J. Virol. 2014, 88, 7904–7914. [Google Scholar] [CrossRef] [PubMed]
- Lingappa, J.R.; Dooher, J.E.; Newman, M.A.; Kiser, P.K.; Klein, K.C. Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 Gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly. J. Biol. Chem. 2006, 281, 3773–3784. [Google Scholar] [CrossRef] [PubMed]
- Robinson, B.A.; Reed, J.C.; Geary, C.D.; Swain, J.V.; Lingappa, J.R. A temporospatial map that defines specific steps at which critical surfaces in the Gag MA and CA domains act during immature HIV-1 capsid assembly in cells. J. Virol. 2014, 88, 5718–5741. [Google Scholar] [CrossRef] [PubMed]
- Mouland, A.J.; Mercier, J.; Luo, M.; Bernier, L.; DesGroseillers, L.; Cohen, E.A. The double-stranded RNA-binding protein Staufen is incorporated in human immunodeficiency virus type 1: Evidence for a role in genomic RNA encapsidation. J. Virol. 2000, 74, 5441–5451. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.C.; Molter, B.; Geary, C.D.; McNevin, J.; McElrath, J.; Giri, S.; Klein, K.C.; Lingappa, J.R. HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly. J. Cell Biol. 2012, 198, 439–456. [Google Scholar] [CrossRef] [PubMed]
- Chatel-Chaix, L.; Clément, J.-F.; Martel, C.; Bériault, V.; Gatignol, A.; DesGroseillers, L.; Mouland, A.J. Identification of Staufen in the human immunodeficiency virus type 1 Gag ribonucleoprotein complex and a role in generating infectious viral particles. Mol. Cell. Biol. 2004, 24, 2637–2648. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, M.; Freed, E.O. New insights into HIV assembly and trafficking. Physiology 2011, 26, 236–251. [Google Scholar] [CrossRef] [PubMed]
- Grigorov, B.; Arcanger, F.; Roingeard, P.; Darlix, J.-L.; Muriaux, D. Assembly of infectious HIV-1 in human epithelial and T-lymphoblastic cell lines. J. Mol. Biol. 2006, 359, 848–862. [Google Scholar] [CrossRef] [PubMed]
- Perlman, M.; Resh, M.D. Identification of an intracellular trafficking and assembly pathway for HIV-1 Gag. Traffic 2006, 7, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Li, H.; Derdowski, A.; Ding, L.; Burnett, A.; Chen, X.; Peters, T.R.; Dermody, T.S.; Woodruff, E.; Wang, J.-J.; et al. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 2005, 120, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Welsch, S.; Müller, B.; Kräusslich, H.-G. More than one door—Budding of enveloped viruses through cellular membranes. FEBS Lett. 2007, 581, 2089–2097. [Google Scholar] [CrossRef] [PubMed]
- Jouvenet, N.; Simon, S.M.; Bieniasz, P.D. Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc. Natl. Acad. Sci. USA 2009, 106, 19114–19119. [Google Scholar] [CrossRef] [PubMed]
- Sardo, L.; Hatch, S.C.; Chen, J.; Nikolaitchik, O.; Burdick, R.C.; Chen, D.; Westlake, C.J.; Lockett, S.; Pathak, V.K.; Hu, W.-S. Dynamics of HIV-1 RNA Near the Plasma Membrane during Virus Assembly. J. Virol. 2015, 89, 10832–10840. [Google Scholar] [CrossRef] [PubMed]
- Bell, N.M.; Lever, A.M.L. HIV Gag polyprotein: Processing and early viral particle assembly. Trends Microbiol. 2013, 21, 136–144. [Google Scholar] [CrossRef] [PubMed]
- Könnyű, B.; Sadiq, S.K.; Turányi, T.; Hírmondó, R.; Müller, B.; Kräusslich, H.-G.; Coveney, P.V.; Müller, V. Gag-Pol processing during HIV-1 virion maturation: A systems biology approach. PLoS Comput. Biol. 2013, 9, e1003103. [Google Scholar] [CrossRef] [PubMed]
- Mattei, S.; Anders, M.; Konvalinka, J.; Kräusslich, H.-G.; Briggs, J.A.G.; Müller, B. Induced maturation of human immunodeficiency virus. J. Virol. 2014, 88, 13722–13731. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, A.H.; Zack, J.A.; Knigge, M.; Paul, D.A.; Kempf, D.J.; Norbeck, D.W.; Swanstrom, R. Partial inhibition of the human immunodeficiency virus type 1 protease results in aberrant virus assembly and the formation of noninfectious particles. J. Virol. 1993, 67, 4050–4055. [Google Scholar] [PubMed]
- Müller, B.; Anders, M.; Akiyama, H.; Welsch, S.; Glass, B.; Nikovics, K.; Clavel, F.; Tervo, H.-M.; Keppler, O.T.; Kräusslich, H.-G. HIV-1 Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. J. Biol. Chem. 2009, 284, 29692–29703. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Rein, A. Maturation of dimeric viral RNA of Moloney murine leukemia virus. J. Virol. 1993, 67, 5443–5449. [Google Scholar] [PubMed]
- Grohman, J.K.; Gorelick, R.J.; Kottegoda, S.; Allbritton, N.L.; Rein, A.; Weeks, K.M. An immature retroviral RNA genome resembles a kinetically trapped intermediate state. J. Virol. 2014, 88, 6061–6068. [Google Scholar] [CrossRef] [PubMed]
- Fu, W.; Gorelick, R.J.; Rein, A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J. Virol. 1994, 68, 5013–5018. [Google Scholar] [PubMed]
- Fu, W.; Dang, Q.; Nagashima, K.; Freed, E.O.; Pathak, V.K.; Hu, W.-S. Effects of Gag mutation and processing on retroviral dimeric RNA maturation. J. Virol. 2006, 80, 1242–1249. [Google Scholar] [CrossRef] [PubMed]
- Levin, J.G.; Guo, J.; Rouzina, I.; Musier-Forsyth, K. Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: Critical role in reverse transcription and molecular mechanism. Prog. Nucleic Acid Res. Mol. Biol. 2005, 80, 217–286. [Google Scholar] [PubMed]
- Ohishi, M.; Nakano, T.; Sakuragi, S.; Shioda, T.; Sano, K.; Sakuragi, J. The relationship between HIV-1 genome RNA dimerization, virion maturation and infectivity. Nucleic Acids Res. 2011, 39, 3404–3417. [Google Scholar] [CrossRef] [PubMed]
- Rein, A.; Henderson, L.E.; Levin, J.G. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: Significance for viral replication. Trends Biochem. Sci. 1998, 23, 297–301. [Google Scholar] [CrossRef]
- De Marco, A.; Müller, B.; Glass, B.; Riches, J.D.; Kräusslich, H.-G.; Briggs, J.A.G. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS Pathog. 2010, 6, e1001215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Naiyer, N.; Mitra, M.; Li, J.; Williams, M.C.; Rouzina, I.; Gorelick, R.J.; Wu, Z.; Musier-Forsyth, K. Distinct nucleic acid interaction properties of HIV-1 nucleocapsid protein precursor NCp15 explain reduced viral infectivity. Nucleic Acids Res. 2014, 42, 7145–7159. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Gorelick, R.J.; Levin, J.G. Selection of fully processed HIV-1 nucleocapsid protein is required for optimal nucleic acid chaperone activity in reverse transcription. Virus Res. 2014, 193, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.; Datta, S.A.K.; Mitra, M.; Gorelick, R.J.; Rein, A.; Levin, J.G. Fundamental differences between the nucleic acid chaperone activities of HIV-1 nucleocapsid protein and Gag or Gag-derived proteins: Biological implications. Virology 2010, 405, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Saadatmand, J.; Niu, M.; Kleiman, L. Roles of Gag and NCp7 in facilitating tRNA(Lys)(3) Annealing to viral RNA in human immunodeficiency virus type 1. J. Virol. 2009, 83, 8099–8107. [Google Scholar] [CrossRef] [PubMed]
- Seif, E.; Niu, M.; Kleiman, L. In virio SHAPE analysis of tRNA(Lys3) annealing to HIV-1 genomic RNA in wild type and protease-deficient virus. Retrovirology 2015, 12, 40. [Google Scholar] [CrossRef] [PubMed]
- Lapadat-Tapolsky, M.; Pernelle, C.; Borie, C.; Darlix, J.L. Analysis of the nucleic acid annealing activities of nucleocapsid protein from HIV-1. Nucleic Acids Res. 1995, 23, 2434–2441. [Google Scholar] [CrossRef] [PubMed]
- Vo, M.-N.; Barany, G.; Rouzina, I.; Musier-Forsyth, K. HIV-1 nucleocapsid protein switches the pathway of transactivation response element RNA/DNA annealing from loop-loop “kissing” to “zipper”. J. Mol. Biol. 2009, 386, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Belfetmi, A.; Zargarian, L.; Tisné, C.; Sleiman, D.; Morellet, N.; Lescop, E.; Maskri, O.; René, B.; Mély, Y.; Fossé, P.; et al. Insights into the mechanisms of RNA secondary structure destabilization by the HIV-1 nucleocapsid protein. RNA 2016, 22, 506–517. [Google Scholar] [CrossRef] [PubMed]
- Didierlaurent, L.; Houzet, L.; Morichaud, Z.; Darlix, J.-L.; Mougel, M. The conserved N-terminal basic residues and zinc-finger motifs of HIV-1 nucleocapsid restrict the viral cDNA synthesis during virus formation and maturation. Nucleic Acids Res. 2008, 36, 4745–4753. [Google Scholar] [CrossRef] [PubMed]
- Houzet, L.; Morichaud, Z.; Didierlaurent, L.; Muriaux, D.; Darlix, J.-L.; Mougel, M. Nucleocapsid mutations turn HIV-1 into a DNA-containing virus. Nucleic Acids Res. 2008, 36, 2311–2319. [Google Scholar] [CrossRef] [PubMed]
- Racine, P.-J.; Chamontin, C.; de Rocquigny, H.; Bernacchi, S.; Paillart, J.-C.; Mougel, M. Requirements for nucleocapsid-mediated regulation of reverse transcription during the late steps of HIV-1 assembly. Sci. Rep. 2016, 6, 27536. [Google Scholar] [CrossRef] [PubMed]
- McCauley, M.J.; Rouzina, I.; Manthei, K.A.; Gorelick, R.J.; Musier-Forsyth, K.; Williams, M.C. Targeted binding of nucleocapsid protein transforms the folding landscape of HIV-1 TAR RNA. Proc. Natl. Acad. Sci. USA 2015, 112, 13555–13560. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Rodríguez, L.; Tsuchihashi, Z.; Fuentes, G.M.; Bambara, R.A.; Fay, P.J. Influence of human immunodeficiency virus nucleocapsid protein on synthesis and strand transfer by the reverse transcriptase in vitro. J. Biol. Chem. 1995, 270, 15005–15011. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Rong, L.; Cherry, E.; Kleiman, L.; Laughrea, M.; Wainberg, M.A. Deletion mutagenesis within the dimerization initiation site of human immunodeficiency virus type 1 results in delayed processing of the p2 peptide from precursor proteins. J. Virol. 1999, 73, 6147–6151. [Google Scholar] [PubMed]
- L’Hernault, A.; Weiss, E.U.; Greatorex, J.S.; Lever, A.M. HIV-2 genome dimerization is required for the correct processing of Gag: A second-site reversion in matrix can restore both processes in dimerization-impaired mutant viruses. J. Virol. 2012, 86, 5867–5876. [Google Scholar] [CrossRef] [PubMed]



© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mailler, E.; Bernacchi, S.; Marquet, R.; Paillart, J.-C.; Vivet-Boudou, V.; Smyth, R.P. The Life-Cycle of the HIV-1 Gag–RNA Complex. Viruses 2016, 8, 248. https://doi.org/10.3390/v8090248
Mailler E, Bernacchi S, Marquet R, Paillart J-C, Vivet-Boudou V, Smyth RP. The Life-Cycle of the HIV-1 Gag–RNA Complex. Viruses. 2016; 8(9):248. https://doi.org/10.3390/v8090248
Chicago/Turabian StyleMailler, Elodie, Serena Bernacchi, Roland Marquet, Jean-Christophe Paillart, Valérie Vivet-Boudou, and Redmond P. Smyth. 2016. "The Life-Cycle of the HIV-1 Gag–RNA Complex" Viruses 8, no. 9: 248. https://doi.org/10.3390/v8090248