3.1. Phylogenetic Relationships of Coccolithoviruses
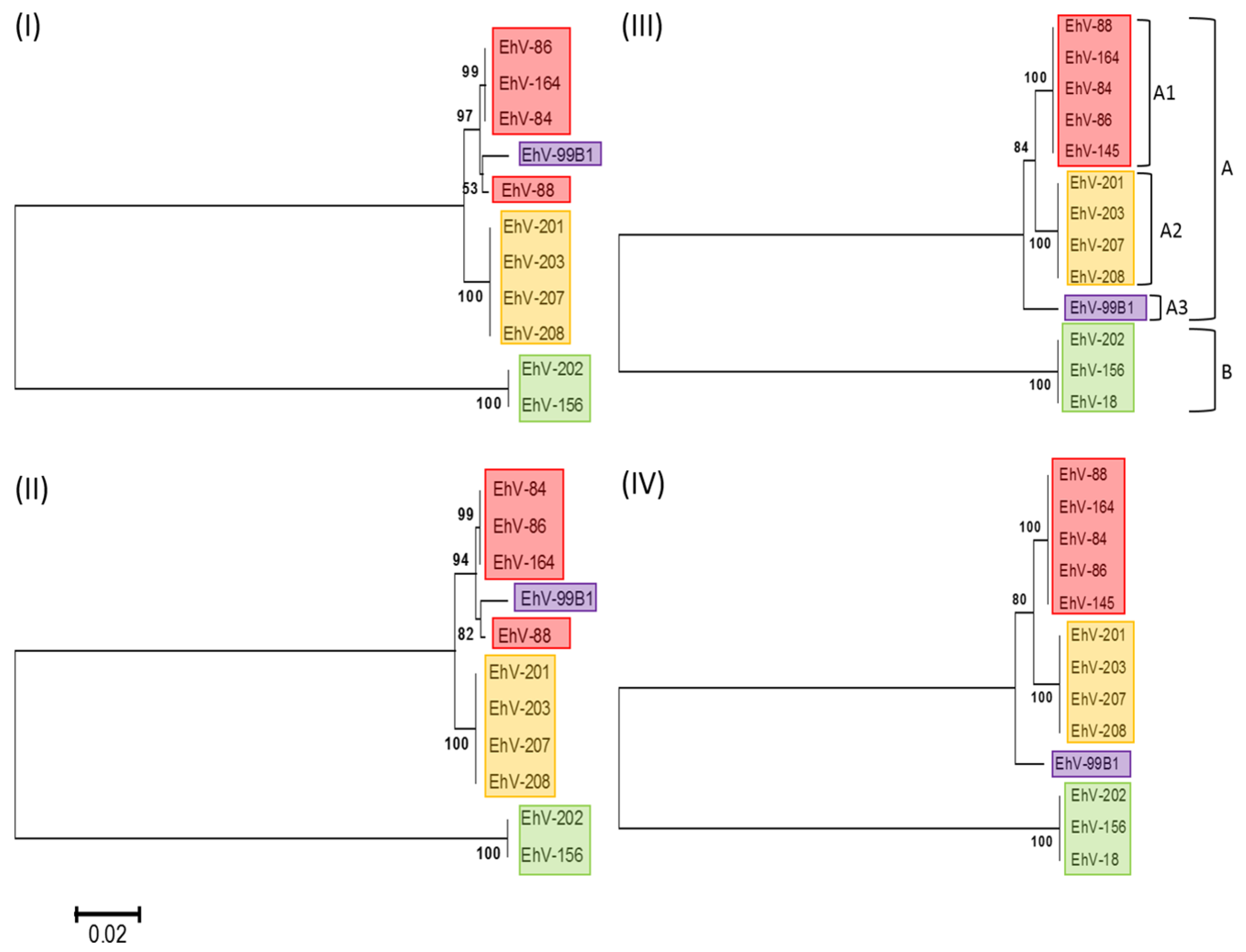
Phylogenetic analysis of available DNA polymerase gene sequences, one of the commonly used phylogenetic markers for the dsDNA viral kingdom, indicated that the EhVs cluster into two major groups: A and B (
Figure 1). The division into these groups was primarily due to a longer DNA polymerase gene present in EhV-18, EhV-156 and EhV-202 (caused by an insert of 12 bp, which results in the addition of glycine, threonine and two prolines at the end of the coding region). Group A was divided into three sub-clusters, where cluster A1 represents strains isolated primarily from the English Channel in 1999, A2 represents strains isolated primarily from the English Channel in 2001, and A3 represents a single strain isolated in 1999 from a Norwegian fjord. As such, these sub-clusters appear to be correlated with both year of isolation and location of origin. However, several exceptions were identified: EhV-145 and EhV-164 also clustered with A1 but were isolated after 1999. EhV-202, isolated in 2001 from the English Channel, clustered in B, with EhV-156 and EhV-18 (due to the 12 bp insertion mentioned previously), rather than A2. To further test these EhV groupings, phylogenetic analysis was also performed using the less conserved serine palmitoyltransferase (SPT) gene. This revealed similar groupings, with the exception of EhV-99B1 which clustered with A1.
The phylogenetic clades were also confirmed by the ANI analysis, in which the closest relative to EhV-86 based on the bidirectional pairwise alignment of the total number of genes was EhV-164 (placed in sub-clade A1), whereas those the furthest away were EhVs in clade B (
Table 2).
Interestingly, significant differences have been observed in the infection dynamics of representative EhVs belonging to each phylogenetic clade identified in this study (
Figure 1), both in respect to their lytic period [
13] and their ability to utilize their host for the production of virally encoded glycosphingolipids, which are crucial for the successful demise of host cells [
67].
3.2. Homology, Heterology and Genome Structure
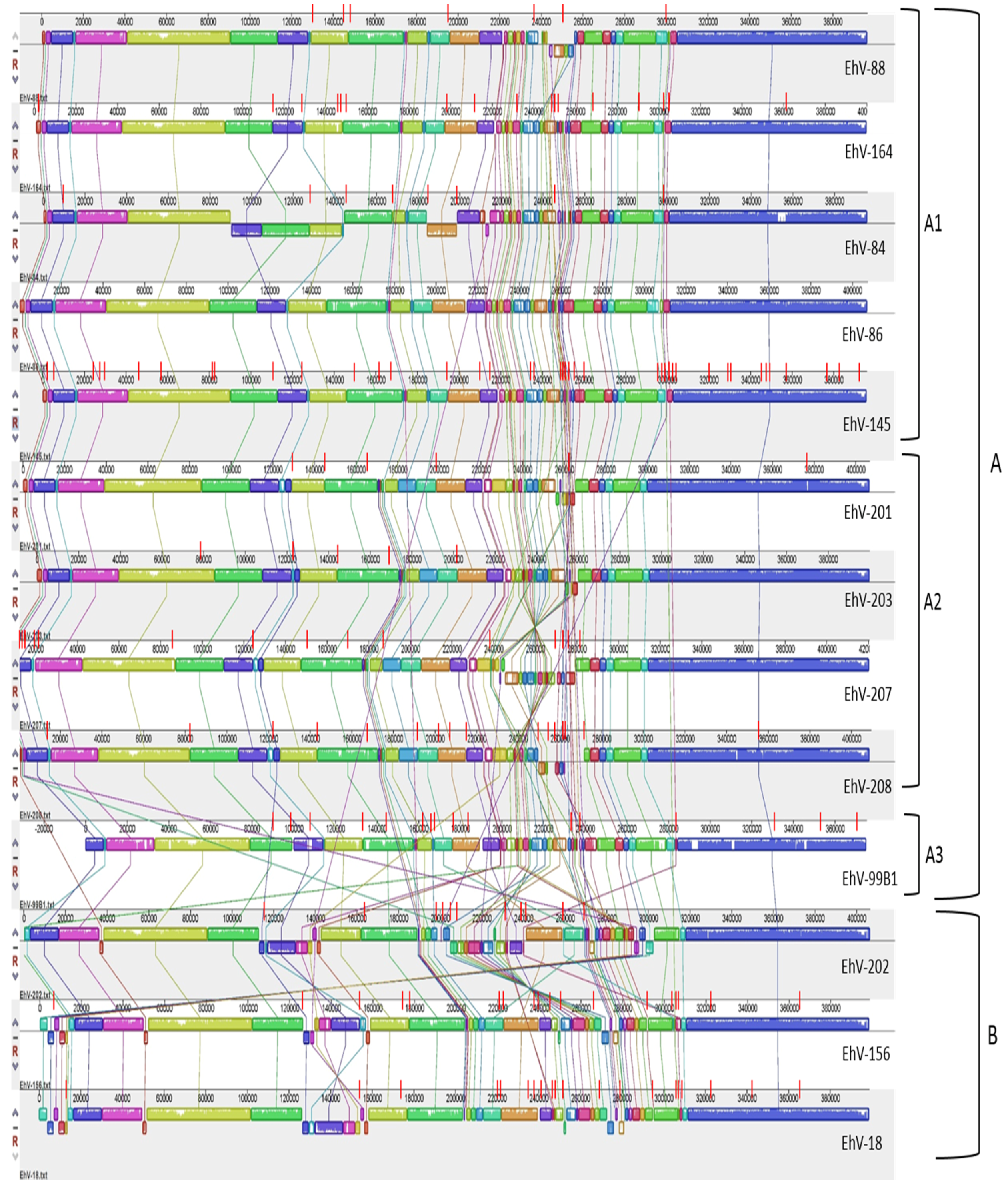
Whole genome alignment revealed that the EhV genomes were syntenic with the exception of a ~80,000 bp section located in the middle of each genome (
Figure 2) enriched with early transcription promoter elements [
1,
20,
68]. As expected [
37], this hyper-variable region was characterized by numerous inversions, rearrangements and base pair substitutions. One possibility is that the observed variation in this region is a result of the gaps in the draft genomes between the different contigs (
Figure 2). Although this may be the case for some of the draft genomes (i.e., EhV-18, EhV-156, EhV-202, and EhV-208), where the highest number of gaps are in that region, it is not for others. For example, EhV-201 and EhV-99B1 sequences only have 2–3 gaps in that region, yet still exhibit variability. Further, many gaps are located in areas outside this region and show little variation; the draft genome of EhV-145 has 10 gaps in the section that ranges from ~300,000 bp to ~397,000 bp, yet variation in this section is minimal. It is therefore likely that the early transcriptional profile of these viruses can differ considerably.
The misalignment of different sections within the hyper-variable region (evident by numerous crossing lines between the co-linear blocks in
Figure 2) becomes more prominent the further a genome is from those in cluster A1 (
Figure 2). This is consistent with the DNA polymerase based phylogeny (
Figure 1), in which viruses belonging to group B are more distant from those in group A1. Considered together, the genome alignments (
Figure 2), and ANI analysis (
Table 2) support the clustering of EhVs into the aforementioned groups and is consistent with DNA polymerase and SPT based phylogenies.
3.3. Genome Size and Putative Gene Differences
Previous pulsed-field gel electrophoresis (PFGE) of coccolithovirus genomes indicates that they have an average size of 410 kb [
2]. In our study, the estimated length of the EhV genomes ranged from 376,759 to 421,891 bp (with a GC content of 39.94% to 40.49%), suggesting considerable variability in EhV genome size. For example, EhV-99B1 appears significantly shorter than the rest of the genomes (
Figure 2). The genomes also differed in the number of predicted protein coding sequences (CDSs). They contained between 444 and 548 CDSs, from which an average of 90 had functional prediction, including three to six transfer RNA (tRNA) genes (
Table 3). Due to the highly repetitive nature of the hyper-variable region, some of the variations observed could be attributed to sequencing errors, misalignments introduced during the original genome assembly, and the numerous gaps between the different contigs. However, these variations may also reflect real differences, as it is unlikely that regions as big as 45,132 bp long (the difference between the longest and shortest genome) were incorrectly sequenced or not included in the assemblies. These variations may have real consequences on the virus life cycle, affecting the time for viral genome synthesis within infected cells and, to an extent, the size of the capsid into which the DNA will be packed. As viruses exploit the resources of the host cell to synthesize new, infectious viral particles, these inherent differences may ultimately affect the number of viable viruses produced during infection.
Although strain-specific differences in the number and composition of transfer RNAs (tRNAs) encoded within the genomes were identified, the tRNAs for Arg, Asn and Gln were common to all (
Table 4). Similarly, Pagarete et al. [
44] report differences in the type of tRNAs encoded by EhV-86 and EhV-99B1. However, after an in-depth study of the different codon frequencies of the genomes of EhV-86 and EhV-99B1, they concluded that the difference in tRNAs does not correspond to the respective codon frequencies in each genome and that it is more likely that strain-specific tRNA differences represent adaptations to different host strains. If so, viruses with a larger number of tRNAs may have an advantage in terms of host range and translation within different host genotypes. Allen et al. [
20] observe (in the absence of statistically robust data) that out of 23 different host strains tested, EhV-207 (which codes for six tRNAs) infects 10, whereas EhV-86 (which codes for five tRNAs) infects eight. Moreover, EhV-207 outcompetes EhV-86 when placed in direct competition over a single host genotype and has a higher rate of virus particle production [
13]. The extent to which tRNAs are responsible for these observations has yet to be determined.
Most predicted CDSs were shared between all EhV genomes but a number of strain-specific CDSs were detected (
Table 5). EhV-84 and EhV-202 contain the largest number of potentially “unique” genes (27 and 18, respectively). This is intriguing, as the other genomes in clade A1 and B (to which EhV-84 and EhV-202 belong respectively) do not exhibit a large number of “unique” genes (
Table 5).
The observed strain-specific genomic differences within members of the same clade (
Table 3,
Table 4 and
Table 5) highlight the limitations of virus taxonomy using a single gene such as DNA polymerase, when one tries to infer upon the evolutionary connections of these large viruses, the origin of certain genes, and potentially their functional implications. These limitations also become apparent in large-scale phylogenetic studies that include numerous taxa across the domains of life. Almost two decades ago, Villarreal and DeFilippis [
69] suggested an alternative hypothesis for the flow of genetic material, with an emphasis on the virus DNA polymerase. Based on a large number of hits of virus DNA polymerases to other taxa, they suggest that algal viruses did not acquire replication genes from their eukaryotic hosts but vice versa [
69]. We performed a similar analysis using the DNA polymerase gene of EhV-86 against the non-redundant protein sequence database in NCBI. The closest hit was to a DNA polymerase of the Yellowstone lake phycodnavirus 1 (reference sequence: YP_009174732.1), with a query cover of 81%, E-value of 9 × 10
−137 and identity of 35% (
Table S1), other
Phycodnaviruses, and other organisms such as
Klebsormidium laccidum (which belongs to a genus of filamentous charophyte green algae).
3.4. Coccolithovirus Gene Similarities to the Emiliania huxleyi Host
The prevailing dogma is that many EhV genes were acquired from
Emiliania huxleyi through HGT. BLASTp analysis of EhV genomes revealed that, on average, ~25 EhV protein coding sequences were highly similar to counterparts in the host genome of
E. huxleyi CCMP1516 (consistent with previous analysis of this host genome [
66]). Unfortunately, most of these genes have no assigned function yet. Those with a predicted function include a group of genes that encode a de novo sphingolipid biosynthesis pathway (i.e., serine palmitoyltransferase [45%], sterol desaturase [42%], transmembrane fatty acid elongation protein [56%], and lipid phosphate phosphatase [29%]). BLASTp analysis of the EhV serine palmitoyltransferase gene (which encodes the rate-limiting enzyme in this pathway) showed that the most similar genes in the non-redundant protein sequence database of NCBI are putative serine palmitoyltransferases in
E. huxleyi,
Chrysochromulina sp. and
Perkinsus marinus (
Table S2). These genes are essential for the progression of infection through the production of virus-derived glycosphingolipids [
28] and induction of PCD pathways within infected cells [
17], and are present in all EhV genomes analysed. As previously reported [
49], the two protein subunits of SPT (LCB1 and LCB2) are encoded by two separate genes in the genomes of EhV-18 and EhV-145. This separation over two genes may reflect the ancestral form prior to the fusion of the two subunits into a single gene (as seen in all other EhVs). The functional implications of this during infection are so far unknown. Nevertheless, the acquisition of this near complete pathway represents a classical example of HGT of a virus with its host, and illustrates a finely tuned co-evolutionary relationship between a host and its virus. In this case, it is manifested by a control for sphingolipid biosynthesis [
29].
With the exception of EhV-99B1, EhV-202, EhV-18 and EhV-156, the CDS with the highest identity (i.e., 86.4%) to a similar gene in
E. huxleyi CCMP1516 is a putative phosphate permease transporter (denoted as
ehv117 in EhV-86). This transporter is encoded by all EhV isolates from the English Channel and the Scottish coast, but absent in the Norwegian isolate EhV-99B1 ([
44]; and this study) and the partially sequenced genome of the Norwegian isolate EhV-163 (not included in this study) [
43]. Instead, a 75-bp scar remnant of the transporter gene is still present at the 3′ end of this gene in both Norwegian isolates, and is replaced by a putative endonuclease [
43]. This indicates that the Norwegian fjord EhVs once possessed the gene [
44] and have since lost it. Such genes are common in
E. huxleyi, which has six inorganic phosphate transporters and an alkaline phosphatase that enable it to thrive in low phosphorous conditions [
66]. Phosphorus is also essential for successful viral replication in most host-virus systems [
70,
71,
72]. However, we do not currently know if the
ehv117 gene product is used to enhance phosphate acquisition from the external environment during infection. Its replacement with an endonuclease encoding gene in EhV-99B1 is intriguing, as this may function to provide additional phosphate via utilization of internal cellular resources, rather than acquisition of external phosphate via transporter activity. This could reflect the selective pressure imposed by particular geographic locations that are characterized by specific physicochemical conditions. For instance, the frequency of phosphate uptake genes in metagenomic datasets is higher in locations where the average phosphate concentration is lower (e.g., North Atlantic Gyre) [
42,
73]. Like SPT, it is most similar to a homologue in
E. huxleyi (
Table S3) and was likely acquired by HGT with its host. Other high hits in NCBI include similar genes in
Ostreococcus lucimarinus and
Ectocarpus siliculosus. Similarly, a phosphate transporter gene (i.e.,
pho4) was identified in the viruses infecting
Ostreococcus and it was proposed that this was also a result of HGT [
42].
For EhVs where the
ehv117 transporter gene is absent, the CDS with the highest percentage similarity to a CDS in
E. huxleyi CCMP1516 varies. In EhV-99B1, it is a predicted ribonucleoside-diphosphate reductase protein (RNR) (71.9% similar) whereas in EhV-202, EhV-18, and EhV-156 it is a predicted polyubiquitin protein (94.7% similar). While the former is an enzyme crucial for the conversion of ribonucleotides to deoxyribonucleotides for DNA synthesis, the latter is involved in protein degradation. We cannot confirm if these genes are also a result of HGT with their host, as there were many top hits in the NCBI non-redundant protein sequence database to other taxa, including the haptophyte
Chrysochromulina,
Chitinophaga niabensis bacteria, and the Gram-negative bacteria
Cnuella takakiae, in the case of RNR (
Table S4), and radiolarians such as
Collozoum inerme, and protists such as
Vitrella brassicaformis, in the case of polyubiquitin (
Table S5).
3.5. Coccolithovirus Gene Similarities to Other Eukaryotes
As previously mentioned, the current consensus is that many EhV genes are a result of HGT between coccolithoviruses and their specific host [
39]. If this is true, it demonstrates that these viruses are entwined within the molecular workings of their hosts and have managed to acquire genes that target and coerce core aspects of their cellular metabolism. Alternatively, many EhV genes (including some of those mentioned in
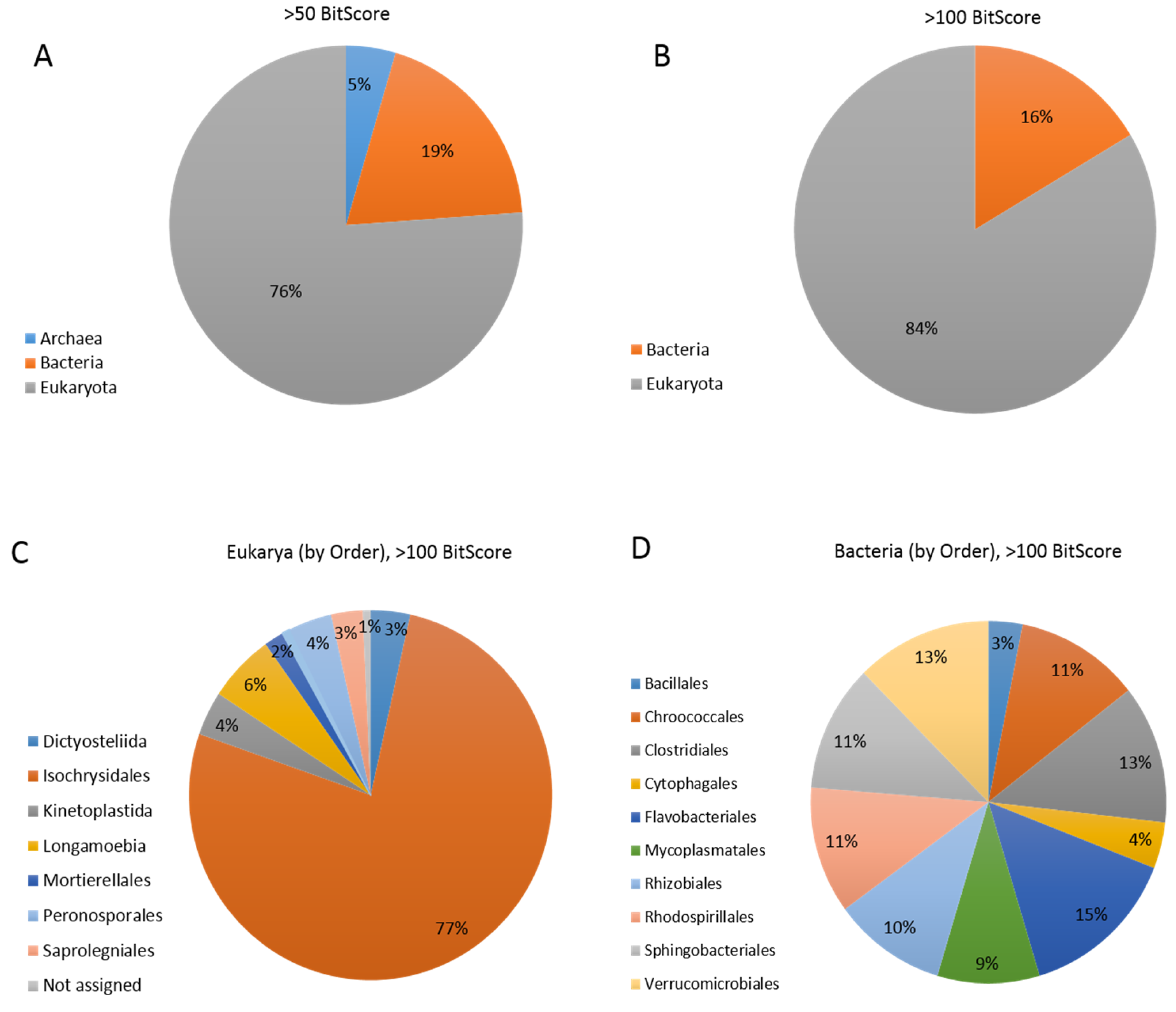
Section 3.4) may have an alternative origin. Thirty-three percent of the total EhV gene hits in the “Best BLAST hit” analysis (which excluded viruses) matched Eukaryotes other than
Isochrysidales (the order to which
E. huxleyi belongs) (
Figure 3). These genes included those involved in nucleotide transport and metabolism, replication, recombination and repair, and transcription (
Table 6). Among these other Eukaryotes are phylogenetically distant organisms such as moulds, fungi, unicellular flagellated protozoa, centric diatoms, amoeba, and green algae. Most of these can be found in the planktonic realm where
E. huxleyi also dwells, particularly diatoms whose development is often tightly linked to
E. huxleyi growth in ecological successions [
74]. In this study, some of the top EhV gene hits against other genera include DNA-directed RNA polymerase subunit-B (an enzyme that produces mainly RNA transcripts) and DNA ligase (an enzyme that facilitates the joining of DNA strands) (
Table 6). The former had top hits against members of the genus
Dictyostelium, a group of soil living amoeba, whereas the latter had top hits against
Micromonas, a group of chlorophyte green algae (
Tables S6 and S7).
It is also plausible that EhVs have, or have had in their evolutionary past, alternative hosts that have enabled them to facilitate HGT with other taxa. Some giant algal viruses have the capacity to alternate between hosts belonging to different genera. For example, viruses capable of infecting
Haptolina hirta infect also
Prymnesium kappa and vice versa; [
75]). Alternatively, there may have been a retroviral stage during the evolution of the
E. huxleyi-virus system. Several EhV genes thought to have originated via HGT with
E. huxleyi (including those for the de novo sphingolipid biosynthesis) lack introns, unlike their counterparts in the host genome. This suggests the possible involvement of coccolithophore-infecting RNA viruses in mediating HGT, although such viruses have not been observed yet.
3.6. Coccolithovirus Gene Similarities to Bacteria
There are close ecological relationships between
E. huxleyi and some bacterial groups. A previous study reports that
E. huxleyi blooms in the North Atlantic are dominated by
Roseobacter, SAR86, SAR11, and other
Alpha and
Gammaproteobacteria [
76]. In their study, bacteria belonging to the
Roseobacter genus and SAR86 and SAR11 clades account, together, for >50% of the bacterial rDNA in surface waters, whereas a cyanobacterium and members of the
Alphaprotobacteria are associated with chlorophyll a-rich waters in the euphotic zone (0–50 m in depth), the typical niche at which coccolithophores are found [
76]. A link between coccolithophores and bacteria is also found in cultures of
E. huxleyi and
Coccolithus pelagicus, which are enriched in hydrocarbon-degrading bacteria belonging to
Marinobacter and
Marivita [
77]. These bacteria can colonize exopolymeric substances exuded by microalgae, but the ecological role of this phenomenon is unknown [
78]. This tight evolutionary coexistence involving
E. huxleyi, its viral community, and associated bacteria may have resulted in HGT across the domains.
In our more conservative “Best BLAST hit” analysis (using a bit-score of >100) we saw that 16% of the EhV genes also shared strong similarities to bacterial genes (
Figure 3), involved in nucleotide, lipid, and carbohydrate transport and metabolism; and replication, recombination and repair (
Table 6). Interestingly, almost half of the bacterial-like genes in the Chlorella virus NY2A, and a fifth of those in Mimivirus were proposed to be involved in DNA replication and repair [
55,
56]. In our analysis, top hits to bacteria include genes that code for sialidase, deoxycytidylate deaminase and fatty acid desaturase. These hits were against members of the genus
Clostridium,
Sphingobacterium and
Niveispirillum respectively (
Table 6, and
Tables S8–S10). Bacteria from these genera are consistently found in all coccolithophore cultures studied by Green et al. [
77]. The hits against the
Sphingobacteriales are particularly intriguing as members of this order are characterized by the presence of cellular lipid components that are comprised of high concentrations of sphingophospholipids [
79]. The rate limiting step in sphingolipid biosynthesis in
S. multivorum is serine palmitoyltransferase (SPT) [
80], as in the
E. huxleyi—virus system [
81]. Although the EhV SPT is more similar to the host gene (as indicated above), the closest crystal structure in the PDB database to the EhV SPT is
Sphingobacterium multivorum [
67]. This structure was used to model the EhV derived SPT enzyme and to infer its catalytic capabilities during infection [
36,
67].
Moreover, in
S. multivorum, sialidase specifically hydrolyzes deaminated neuraminic acid linkages and is localized in the periplasm [
82,
83]. Viruses such as influenza, adenoviruses, and polyomaviruses [
84,
85], can use virus-derived sialidase to target host sialic acid glycosphingolipids located on the host membrane. A similar process could be fundamental for EhV host recognition and entry, notably because susceptible
E. huxleyi strains are enriched in glycosphingolipids (GSLs) with a sialic acid-modified glycosyl head-group [
12,
14]. These GSLs are proposed as a target for hydrolysis by EhV-encoded sialidases or as a ligand for the attachment of EhV lectin proteins [
23]. Indeed, virus-derived sialidase transcripts are detected during EhV-86 infection 6–24 h post infection [
21].
Surprisingly, we observed that the putative sialidase gene is severely truncated (up to 95% of its extension) in some EhV strains. Its full form is present in 1999 isolates (as well as EhV-164 and EhV-145), whereas those isolated in 2001 (as well as in EhV-18 and EhV-156) have a truncated form. In the genomes with the truncated form, the reduction is from a full gene of 1122 bp to 180 bp. Further reduction is seen in EhV-18 and EhV-156 where only a 60 bp fragment of the original gene remains. To date, EhV infectivity data do not show an association between the lack of a full length sialidase gene and a reduced ability to infect
E. huxleyi. Hence, the selection against this gene in many EhVs and the fact that infection still occurs (presumably alongside other viruses able to utilize the active gene), suggests that its presence is not essential. Previous studies of Influenza A viruses show that a lack of functional sialidase is not essential to all viruses and that some viruses can adapt and grow in its absence in tissue cultures, mice, and embryonic eggs [
86].
The role of two other bacterial-like genes, fatty acid desaturase and deoxycytidylate deaminase (
Table 6) identified in EhVs requires further investigation. Fatty acid desaturases catalyze the desaturation of fatty acids. During EhV infection, a significant remodeling of the fatty acid profile of
E. huxleyi occurs and this gene most likely plays an important role [
30,
87]. Deoxycytidylate deaminase is an enzyme involved in the biosynthesis of deoxyribonucleoside triphosphates (dNTPs). Its role, however, during infection remains elusive, as its expression has not been confirmed [
1].
3.7. Possible Mode for HGT with Bacteria
Bacterial homologues in the genomes of nucleocytoplasmic large DNA viruses (NCLDVs) have been previously reported [
55,
56]. To our knowledge, our study is the first time that this is specifically identified in coccolithoviruses. While it is possible that some of the aforementioned genes were incorporated in the EhV genomes before the evolutionary divergence of eukaryotes and prokaryotes (a detailed “Best BLAST hit” analysis against ancestors of
E. huxleyi such as
Tisochrysis lutea and
Prymnesium parvum, as well as against other primitive lineages of bacteria should reveal that), other explanations are also plausible. HGT could have occurred in situations where the genetic material of EhVs and bacteria co-occurred during virus replication. It is possible that EhVs and bacteria share, at moments, the same intracellular environment within
E. huxleyi, particularly in light of recent data highlighting the importance of mixotrophy in phytoplankton [
88]. For instance, in another NCLDV (i.e.,
Cafeteria roenbergensis virus), a 38-kb genomic region was suggested to be bacterial in origin and possibly acquired during an infection of its microflagellated grazer host that often contains a phagocytosed bacteria in its cytoplasm [
89]. While there is no direct evidence for the engulfment of bacteria by
E. huxleyi, a recent study of the different life stages of this alga shows that both diploid calcifying and motile haploid cells can engulf microbeads as big as 0.5 µm in diameter [
90]. Also, a high abundance of transcripts linked to the digestive apparatus and those related to endocytosis are detected in diploid
E. huxleyi cells, suggestive of an inherent ability of coccolithophores to participate in mixotrophy [
90]. Endocytotic vesicles may be an additional mechanism for EhV infection [
22], and
E. huxleyi feeding on cohabiting bacteria might sometimes be infected by lytic viruses. Such mixotrophic-viral infection events may facilitate HGT across the domains.
Whether these events have occurred multiple times during which individual bacterial genes from different co-habiting bacteria were acquired by EhVs throughout their evolutionary history, or there have been only a few events during which large “chunks” of bacterial DNA (comprising of several adjacent genes, possibly sharing a similar function) were incorporated into EhVs, is unknown. However, the EhV genes with the highest bit-scores to Bacteria in our study were not genomically-located near each other in the analyzed genomes and did not appear to hit the same bacterial taxa. This is therefore consistent with the occurrence of multiple independent gene transfer events.